Functionalization of Ruthenium(II)( eta 6 -p-cymene)(3-hydroxy-2-pyridone) Complexes with (Thio)Morpholine: Synthesis and Bioanalytical Studies.
Hanif, M., Meier, S.M., Adhireksan, Z., Henke, H., Martic, S., Movassaghi, S., Labib, M., Kandioller, W., Jamieson, S.M.F., Hejl, M., Jakupec, M.A., Kraatz, H.B., Davey, C.A., Keppler, B.K., Hartinger, C.G.(2017) Chempluschem 82: 841-847
- PubMed: 31961568
- DOI: https://doi.org/10.1002/cplu.201700050
- Primary Citation of Related Structures:
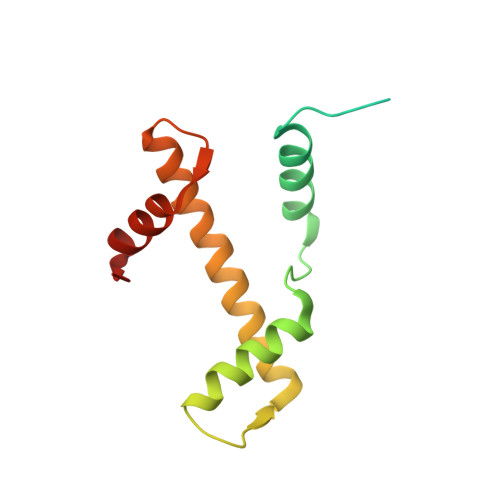
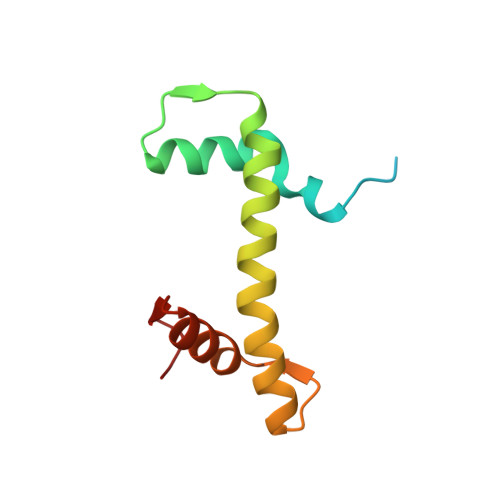
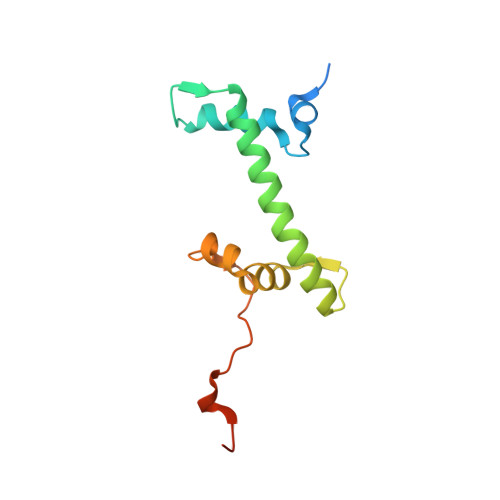
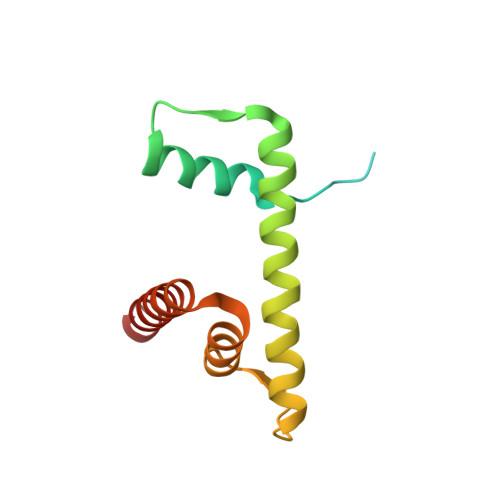
4XUJ - PubMed Abstract:
Hydroxypyr(id)ones constitute an emerging platform for the design of drug molecules, owing to their favorable biocompatibility and toxicity profiles. Herein, [Ru II (η 6 -p-cymene)] complexes with 3-hydroxy-2-pyridinone functionalized with morpholine and thiomorpholine, as a means often used in medicinal chemistry to alter the physicochemical properties of drug compounds, are reported. The compounds underwent hydrolysis of the Ru-Cl bond and the aqua species were stable for up to 48 h in aqueous solution, as observed by 1 H NMR spectroscopy and ESI-MS. The compounds formed adducts with amino acids and proteins through cleavage of the pyridinone ligand. Binding experiments to the nucleosome core particle by means of X-ray crystallography revealed similar reactivity and exclusive binding to histidine moieties of the histone proteins. Preliminary cyclin-dependent kinase 2 (CDK2)/cyclin A kinase inhibitory studies revealed promising activity similar to that of structurally related organometallic compounds.
- School of Chemical Sciences, University of Auckland, Private Bag 92019, Auckland, 1142, New Zealand.
Organizational Affiliation: