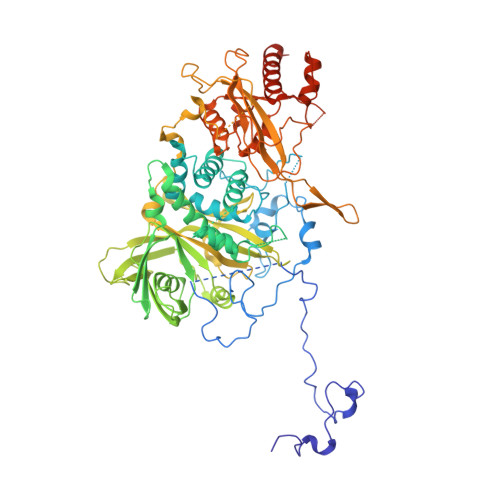
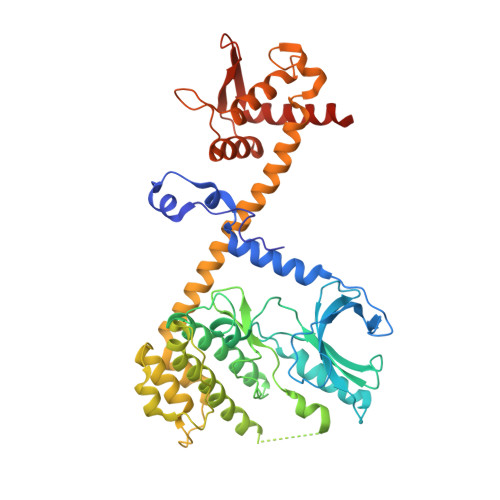
The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase.
Schafer, I.B., Rode, M., Bonneau, F., Schussler, S., Conti, E.(2014) Nat Struct Mol Biol 21: 591-598
- PubMed: 24880344
- DOI: https://doi.org/10.1038/nsmb.2834
- Primary Citation of Related Structures:
4Q8G, 4Q8H, 4XR7 - PubMed Abstract:
Pan2-Pan3 is a conserved complex involved in the shortening of mRNA poly(A) tails, the initial step in eukaryotic mRNA turnover. We show that recombinant Saccharomyces cerevisiae Pan2-Pan3 can deadenylate RNAs in vitro without needing the poly(A)-binding protein Pab1. The crystal structure of an active ~200-kDa core complex reveals that Pan2 and Pan3 interact with an unusual 1:2 stoichiometry imparted by the asymmetric nature of the Pan3 homodimer. An extended region of Pan2 wraps around Pan3 and provides a major anchoring point for complex assembly. A Pan2 module formed by the pseudoubiquitin-hydrolase and RNase domains latches onto the Pan3 pseudokinase with intertwined interactions that orient the deadenylase active site toward the A-binding site of the interacting Pan3. The molecular architecture of Pan2-Pan3 suggests how the nuclease and its pseudokinase regulator act in synergy to promote deadenylation.
- Structural Cell Biology Department, Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: