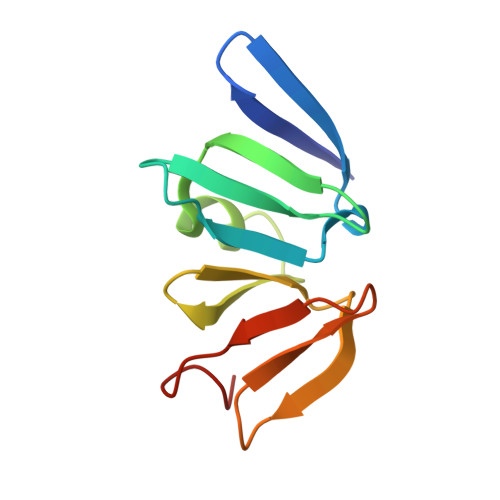
Conformational features of theStaphylococcus aureusAgrA-promoter interactions rationalize quorum-sensing triggered gene expression.
Rajasree, K., Fasim, A., Gopal, B.(2016) Biochem Biophys Rep 6: 124-134
- PubMed: 28955870
- DOI: https://doi.org/10.1016/j.bbrep.2016.03.012
- Primary Citation of Related Structures:
4XQJ, 4XQN, 4XQQ, 4XXE, 4XYO, 4XYQ - PubMed Abstract:
The intracellular trigger for the quorum sensing response mechanism in Staphylococcus aureus involves the phosphorylation of the response regulator AgrA by the membrane anchored histidine kinase AgrC. AgrA activates transcription from three promoter sequences (P1-P3). The promoter strength, conditional association of AgrA with these promoter elements and temporal delay in AgrA-mediated changes in gene expression contribute to the diversity of the quorum sensing response in different S. aureus strains. AgrA promoters comprise of imperfect direct repeats of DNA with a consensus sequence- [TA][AC][CA]GTTN[AG][TG]. Here we describe crystal structures of the DNA-binding (LytTR) domain of AgrA with different cognate DNA sequences that reveal a hitherto unanticipated feature of AgrA-DNA interactions. AgrA promoter interactions are asymmetric with fewer interactions at the binding site proximal to the -35 promoter element. Biochemical assays to evaluate AgrA-promoter interactions suggests that phosphorylation induced dimerization of AgrA can compensate for the asymmetry in AgrA-DNA interactions. The structures also provide a basis to rationalize mutations that were noted to alter AgrA activity without affecting protein-DNA interactions. Put together, the structural data, gene expression and mutational analysis reveal that promoter strength and AgrA phosphorylation enable quorum-sensing triggered transcriptional changes leading to a transition from the persistent to virulent phenotype.
- Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560012, India.
Organizational Affiliation: