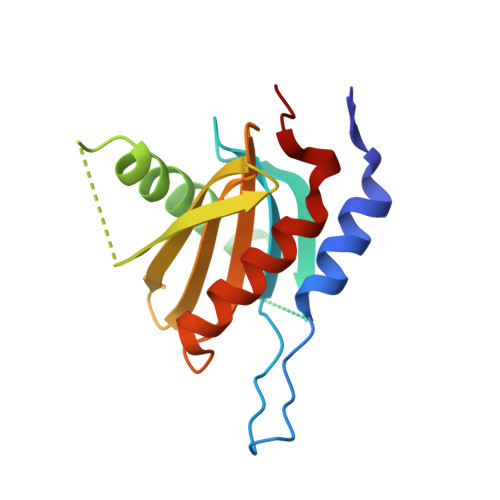
Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling.
Powis, K., Zhang, T., Panchaud, N., Wang, R., De Virgilio, C., Ding, J.(2015) Cell Res 25: 1043-1059
- PubMed: 26206314
- DOI: https://doi.org/10.1038/cr.2015.86
- Primary Citation of Related Structures:
4XPM - PubMed Abstract:
The target of rapamycin complex 1 (TORC1) integrates various hormonal and nutrient signals to regulate cell growth, proliferation, and differentiation. Amino acid-dependent activation of TORC1 is mediated via the yeast EGO complex (EGOC) consisting of Gtr1, Gtr2, Ego1, and Ego3. Here, we identify the previously uncharacterized Ycr075w-a/Ego2 protein as an additional EGOC component that is required for the integrity and localization of the heterodimeric Gtr1-Gtr2 GTPases, equivalent to mammalian Rag GTPases. We also report the crystal structure of the Ego1-Ego2-Ego3 ternary complex (EGO-TC) at 2.4 Å resolution, in which Ego2 and Ego3 form a heterodimer flanked along one side by Ego1. Structural data also reveal the structural conservation of protein components between the yeast EGO-TC and the human Ragulator, which acts as a GEF for Rag GTPases. Interestingly, however, artificial tethering of Gtr1-Gtr2 to the vacuolar membrane is sufficient to activate TORC1 in response to amino acids even in the absence of the EGO-TC. Our structural and functional data therefore support a model in which the EGO-TC acts as a scaffold for Rag GTPases in TORC1 signaling.
- Department of Biology, University of Fribourg, Fribourg CH-1700, Switzerland.
Organizational Affiliation: