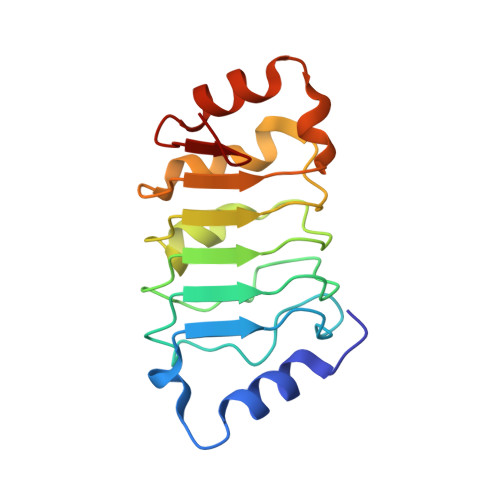
High-resolution crystal structure of the leucine-rich repeat domain of the human tumour suppressor PP32A (ANP32A).
Zamora-Caballero, S., Siauciunaite-Gaubard, L., Bravo, J.(2015) Acta Crystallogr F Struct Biol Commun 71: 684-687
- PubMed: 26057796
- DOI: https://doi.org/10.1107/S2053230X15006457
- Primary Citation of Related Structures:
4XOS - PubMed Abstract:
Acidic leucine-rich nuclear phosphoprotein 32A (PP32A) is a tumour suppressor whose expression is altered in many cancers. It is an apoptotic enhancer that stimulates apoptosome-mediated caspase activation and also forms part of a complex involved in caspase-independent apoptosis (the SET complex). Crystals of a fragment of human PP32A corresponding to the leucine-rich repeat domain, a widespread motif suitable for protein-protein interactions, have been obtained. The structure has been refined to 1.56 Å resolution. This domain was previously solved at 2.4 and 2.69 Å resolution (PDB entries 2je0 and 2je1, respectively). The new high-resolution structure shows some differences from previous models: there is a small displacement in the turn connecting the first α-helix (α1) to the first β-strand (β1), which slightly changes the position of α1 in the structure. The shift in the turn is observed in the context of a new crystal packing unrelated to those of previous structures.
- Department of Genomics and Proteomics, Instituto de Biomedicina de Valencia - CSIC, Jaime Roig 11, 46010 Valencia, Spain.
Organizational Affiliation: