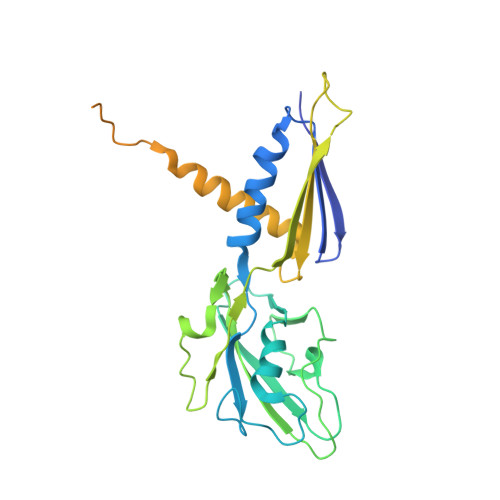
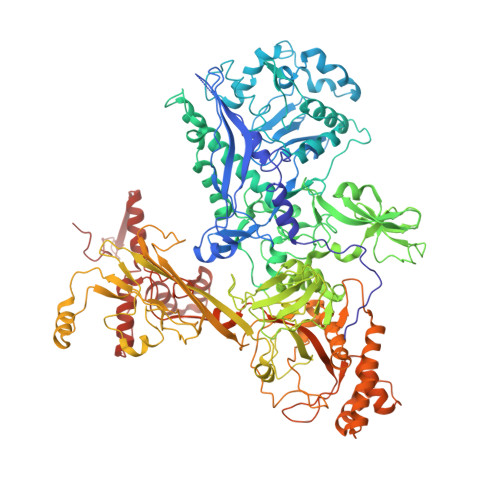
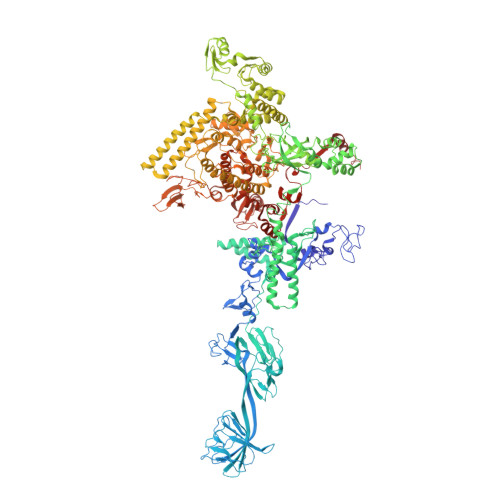
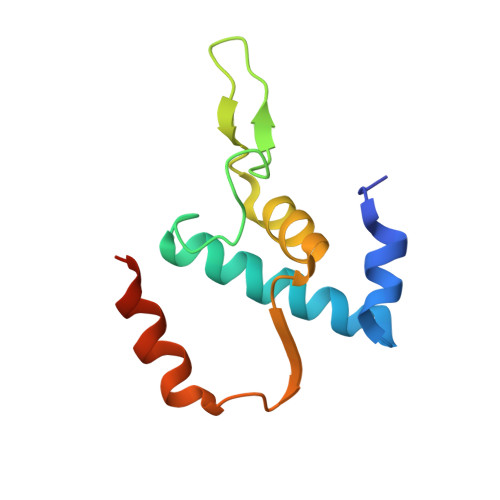
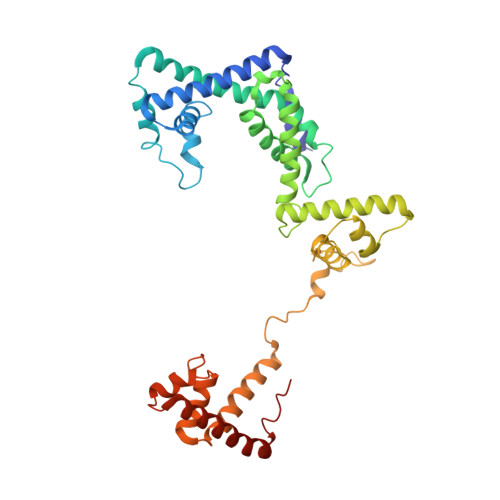
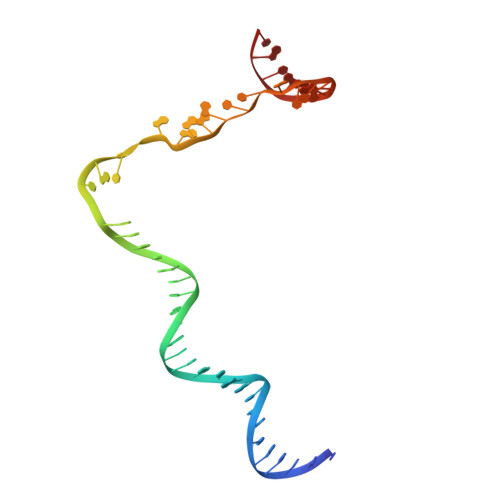
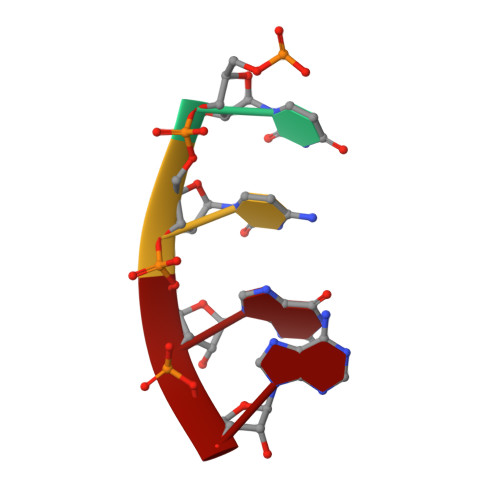
Structure of a bacterial RNA polymerase holoenzyme open promoter complex.
Bae, B., Feklistov, A., Lass-Napiorkowska, A., Landick, R., Darst, S.A.(2015) Elife 4
- PubMed: 26349032
- DOI: https://doi.org/10.7554/eLife.08504
- Primary Citation of Related Structures:
4XLN, 4XLP, 4XLQ - PubMed Abstract:
Initiation of transcription is a primary means for controlling gene expression. In bacteria, the RNA polymerase (RNAP) holoenzyme binds and unwinds promoter DNA, forming the transcription bubble of the open promoter complex (RPo). We have determined crystal structures, refined to 4.14 Å-resolution, of RPo containing Thermus aquaticus RNAP holoenzyme and promoter DNA that includes the full transcription bubble. The structures, combined with biochemical analyses, reveal key features supporting the formation and maintenance of the double-strand/single-strand DNA junction at the upstream edge of the -10 element where bubble formation initiates. The results also reveal RNAP interactions with duplex DNA just upstream of the -10 element and potential protein/DNA interactions that direct the DNA template strand into the RNAP active site. Addition of an RNA primer to yield a 4 base-pair post-translocated RNA:DNA hybrid mimics an initially transcribing complex at the point where steric clash initiates abortive initiation and σ(A) dissociation.
- Laboratory for Molecular Biophysics, The Rockefeller University, New York, United States.
Organizational Affiliation: