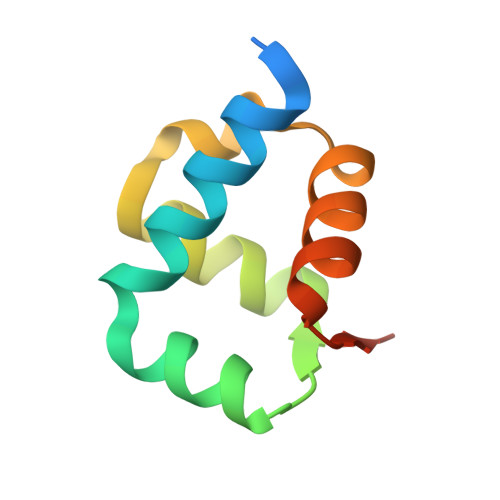
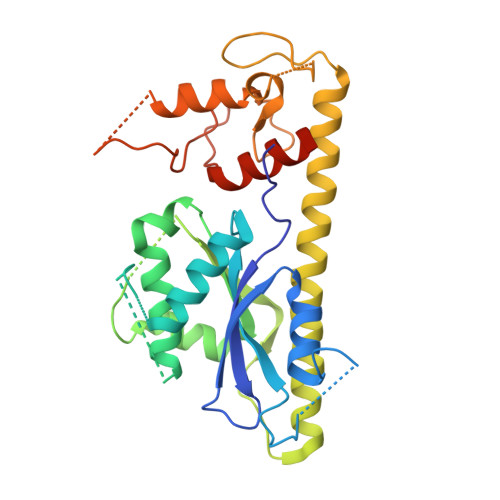
Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Gaur, V., Wyatt, H.D., Komorowska, W., Szczepanowski, R.H., de Sanctis, D., Gorecka, K.M., West, S.C., Nowotny, M.(2015) Cell Rep 10: 1467-1476
- PubMed: 25753413
- DOI: https://doi.org/10.1016/j.celrep.2015.02.019
- Primary Citation of Related Structures:
4XLG, 4XM5 - PubMed Abstract:
The SLX1-SLX4 endonuclease required for homologous recombination and DNA repair in eukaryotic cells cleaves a variety of branched DNA structures. The nuclease subunit SLX1 is activated by association with a scaffolding protein SLX4. At the present time, little is known about the structure of SLX1-SLX4 or its mechanism of action. Here, we report the structural insights into SLX1-SLX4 by detailing the crystal structure of Candida glabrata (Cg) Slx1 alone and in combination with the C-terminal region of Slx4. The structure of Slx1 reveals a compact arrangement of the GIY-YIG nuclease and RING domains, which is reinforced by a long α helix. Slx1 forms a stable homodimer that blocks its active site. Slx1-Slx4 interaction is mutually exclusive with Slx1 homodimerization, suggesting a mechanism for Slx1 activation by Slx4.
- Laboratory of Protein Structure, International Institute of Molecular and Cell Biology, 4 Księcia Trojdena Street, 02-109 Warsaw, Poland.
Organizational Affiliation: