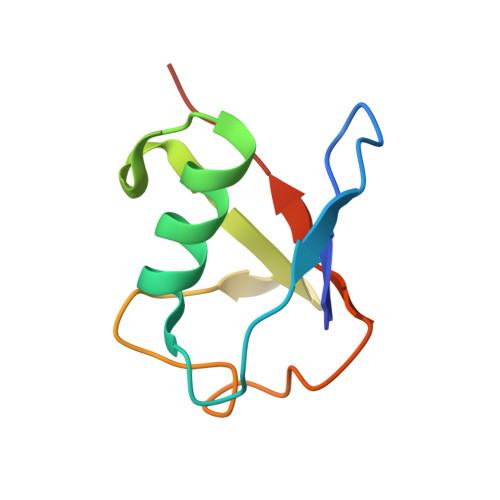
Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains.
Rahighi, S., Braunstein, I., Ternette, N., Kessler, B., Kawasaki, M., Kato, R., Matsui, T., Weiss, T.M., Stanhill, A., Wakatsuki, S.(2016) Structure 24: 412-422
- PubMed: 26876100
- DOI: https://doi.org/10.1016/j.str.2015.12.017
- Primary Citation of Related Structures:
4XKH - PubMed Abstract:
Lys48-linked ubiquitin chains act as the main targeting signals for protein degradation by the proteasome. Here we report selective binding of AIRAPL, a protein that associates with the proteasome upon exposure to arsenite, to Lys48-linked tri-ubiquitin chains. AIRAPL comprises two ubiquitin-interacting motifs in tandem (tUIMs) that are linked through a flexible inter-UIM region. In the complex crystal structure UIM1 binds the proximal ubiquitin, whereas UIM2 (the double-sided UIM) binds non-symmetrically to the middle and distal ubiquitin moieties on either side of the helix. Specificity of AIRAPL for Lys48-linked ubiquitin chains is determined by UIM2, and the flexible inter-UIM linker increases avidity by placing the two UIMs in an orientation that facilitates binding of the third ubiquitin to UIM1. Unlike middle and proximal ubiquitins, distal ubiquitin binds UIM2 through a novel surface, which leaves the Ile44 hydrophobic patch accessible for binding to the proteasomal ubiquitin receptors.
- Structural Biology Research Center, Photon Factory, Institute of Materials Structure Science, High Energy Accelerator Research Organization (KEK), Tsukuba, Ibaraki 305-0801, Japan; Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: