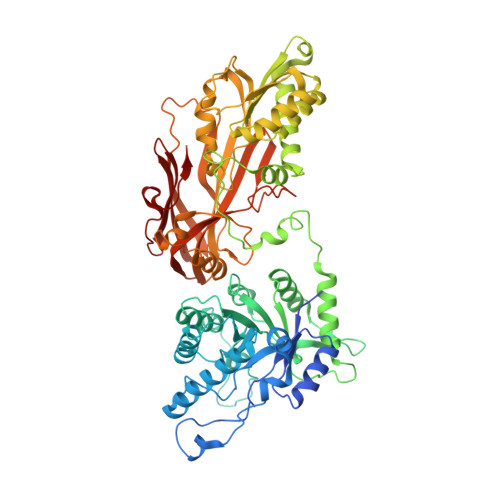
A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Chan-Penebre, E., Kuplast, K.G., Majer, C.R., Boriack-Sjodin, P.A., Wigle, T.J., Johnston, L.D., Rioux, N., Munchhof, M.J., Jin, L., Jacques, S.L., West, K.A., Lingaraj, T., Stickland, K., Ribich, S.A., Raimondi, A., Scott, M.P., Waters, N.J., Pollock, R.M., Smith, J.J., Barbash, O., Pappalardi, M., Ho, T.F., Nurse, K., Oza, K.P., Gallagher, K.T., Kruger, R., Moyer, M.P., Copeland, R.A., Chesworth, R., Duncan, K.W.(2015) Nat Chem Biol 11: 432-437
- PubMed: 25915199
- DOI: https://doi.org/10.1038/nchembio.1810
- Primary Citation of Related Structures:
4X60, 4X61, 4X63 - PubMed Abstract:
Protein arginine methyltransferase-5 (PRMT5) is reported to have a role in diverse cellular processes, including tumorigenesis, and its overexpression is observed in cell lines and primary patient samples derived from lymphomas, particularly mantle cell lymphoma (MCL). Here we describe the identification and characterization of a potent and selective inhibitor of PRMT5 with antiproliferative effects in both in vitro and in vivo models of MCL. EPZ015666 (GSK3235025) is an orally available inhibitor of PRMT5 enzymatic activity in biochemical assays with a half-maximal inhibitory concentration (IC50) of 22 nM and broad selectivity against a panel of other histone methyltransferases. Treatment of MCL cell lines with EPZ015666 led to inhibition of SmD3 methylation and cell death, with IC50 values in the nanomolar range. Oral dosing with EPZ015666 demonstrated dose-dependent antitumor activity in multiple MCL xenograft models. EPZ015666 represents a validated chemical probe for further study of PRMT5 biology and arginine methylation in cancer and other diseases.
- Departments of Biology and Molecular Discovery, Epizyme, Inc., Cambridge, Massachusetts, USA.
Organizational Affiliation: