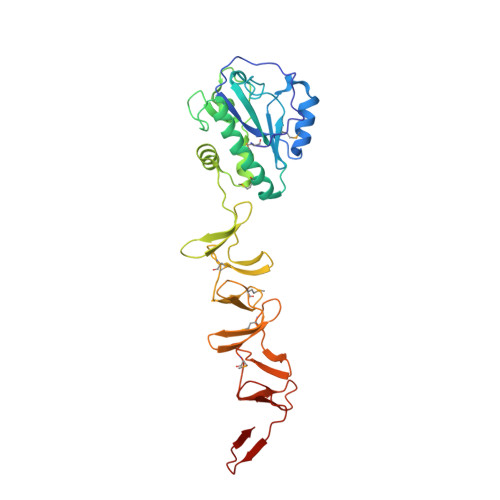
Full-length structure of the major autolysin LytA.
Li, Q., Cheng, W., Morlot, C., Bai, X.H., Jiang, Y.L., Wang, W., Roper, D.I., Vernet, T., Dong, Y.H., Chen, Y., Zhou, C.Z.(2015) Acta Crystallogr D Biol Crystallogr 71: 1373-1381
- PubMed: 26057677
- DOI: https://doi.org/10.1107/S1399004715007403
- Primary Citation of Related Structures:
4X36 - PubMed Abstract:
LytA is responsible for the autolysis of many Streptococcus species, including pathogens such as S. pneumoniae, S. pseudopneumoniae and S. mitis. However, how this major autolysin achieves full activity remains unknown. Here, the full-length structure of the S. pneumoniae LytA dimer is reported at 2.1 Å resolution. Each subunit has an N-terminal amidase domain and a C-terminal choline-binding domain consisting of six choline-binding repeats, which form five canonical and one single-layered choline-binding sites. Site-directed mutageneses combined with enzymatic activity assays indicate that dimerization and binding to choline are two independent requirements for the autolytic activity of LytA in vivo. Altogether, it is suggested that dimerization and full occupancy of all choline-binding sites through binding to choline-containing TA chains enable LytA to adopt a fully active conformation which allows the amidase domain to cleave two lactyl-amide bonds located about 103 Å apart on the peptidoglycan.
- Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China.
Organizational Affiliation: