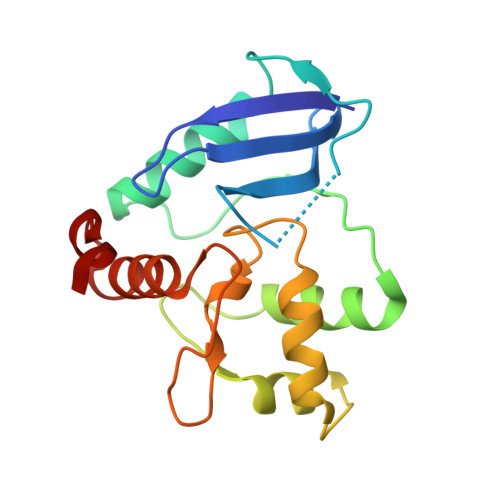
Crystal structure of Mycobacterium tuberculosis O6-methylguanine-DNA methyltransferase protein clusters assembled on to damaged DNA.
Miggiano, R., Perugino, G., Ciaramella, M., Serpe, M., Rejman, D., Pav, O., Pohl, R., Garavaglia, S., Lahiri, S., Rizzi, M., Rossi, F.(2016) Biochem J 473: 123-133
- PubMed: 26512127
- DOI: https://doi.org/10.1042/BJ20150833
- Primary Citation of Related Structures:
4WX9, 4WXC, 4WXD - PubMed Abstract:
Mycobacterium tuberculosis O(6)-methylguanine-DNA methyltransferase (MtOGT) contributes to protect the bacterial GC-rich genome against the pro-mutagenic potential of O(6)-methylated guanine in DNA. Several strains of M. tuberculosis found worldwide encode a point-mutated O(6)-methylguanine-DNA methyltransferase (OGT) variant (MtOGT-R37L), which displays an arginine-to-leucine substitution at position 37 of the poorly functionally characterized N-terminal domain of the protein. Although the impact of this mutation on the MtOGT activity has not yet been proved in vivo, we previously demonstrated that a recombinant MtOGT-R37L variant performs a suboptimal alkylated-DNA repair in vitro, suggesting a direct role for the Arg(37)-bearing region in catalysis. The crystal structure of MtOGT complexed with modified DNA solved in the present study reveals details of the protein-protein and protein-DNA interactions occurring during alkylated-DNA binding, and the protein capability also to host unmodified bases inside the active site, in a fully extrahelical conformation. Our data provide the first experimental picture at the atomic level of a possible mode of assembling three adjacent MtOGT monomers on the same monoalkylated dsDNA molecule, and disclose the conformational flexibility of discrete regions of MtOGT, including the Arg(37)-bearing random coil. This peculiar structural plasticity of MtOGT could be instrumental to proper protein clustering at damaged DNA sites, as well as to protein-DNA complexes disassembling on repair.
- DSF-Dipartimento di Scienze del Farmaco, University of Piemonte Orientale, 28100 Novara, Italy.
Organizational Affiliation: