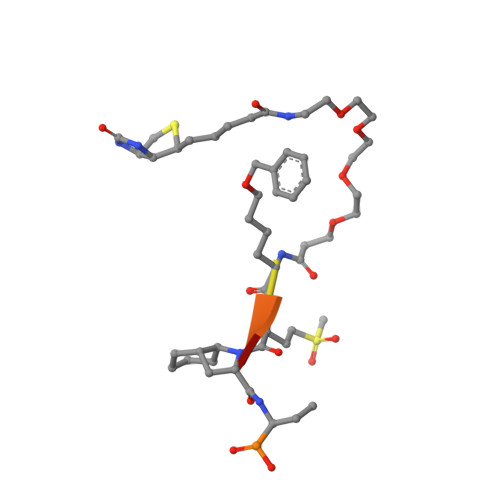
The Elastase-PK101 Structure: Mechanism of an Ultrasensitive Activity-based Probe Revealed.
Lechtenberg, B.C., Kasperkiewicz, P., Robinson, H., Drag, M., Riedl, S.J.(2015) ACS Chem Biol 10: 945-951
- PubMed: 25581168
- DOI: https://doi.org/10.1021/cb500909n
- Primary Citation of Related Structures:
4WVP - PubMed Abstract:
Human neutrophil elastase (HNE) plays a central role in neutrophil host defense, but its broad specificity makes HNE a difficult target for both inhibitor and probe development. Recently, we identified the unnatural amino acid containing activity-based probe PK101, which exhibits astounding sensitivity and selectivity for HNE, yet completely lacks mechanistic explanation for its unique characteristics. Here, we present the crystal structure of the HNE-PK101 complex which not only reveals the basis for PK101 ultrasensitivity but also uncovers so far unrecognized HNE features. Strikingly, the Nle(O-Bzl) function in the P4 position of PK101 reveals and leverages an "exo-pocket" on HNE as a critical factor for selectivity. Furthermore, the PK101 P3 position harbors a methionine dioxide function, which mimics a post-translationally oxidized methionine residue and forms a critical hydrogen bond to the backbone amide of Gly219 of HNE. Gly219 resides in a Gly-Gly motif that is unique to HNE, yet compulsory for this interaction. Consequently, this feature enables HNE to accommodate substrates that have undergone methionine oxidation, which constitutes a hallmark post-translational modification of neutrophil signaling.
- †Sanford-Burnham Medical Research Institute, 10901 North Torrey Pines Road, La Jolla, California 92037, United States.
Organizational Affiliation: