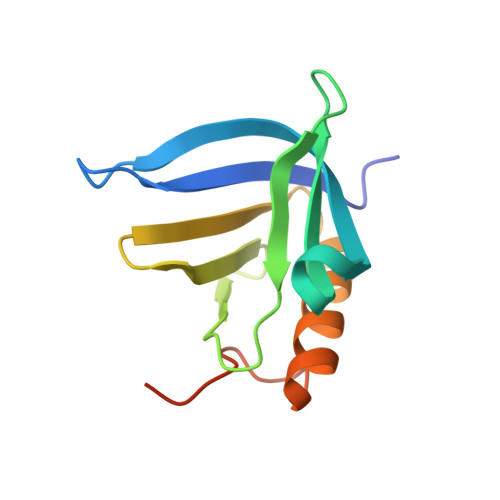
Centromeric binding and activity of Protein Phosphatase 4.
Lipinszki, Z., Lefevre, S., Savoian, M.S., Singleton, M.R., Glover, D.M., Przewloka, M.R.(2015) Nat Commun 6: 5894-5894
- PubMed: 25562660
- DOI: https://doi.org/10.1038/ncomms6894
- Primary Citation of Related Structures:
4WSF - PubMed Abstract:
The cell division cycle requires tight coupling between protein phosphorylation and dephosphorylation. However, understanding the cell cycle roles of multimeric protein phosphatases has been limited by the lack of knowledge of how their diverse regulatory subunits target highly conserved catalytic subunits to their sites of action. Phosphoprotein phosphatase 4 (PP4) has been recently shown to participate in the regulation of cell cycle progression. We now find that the EVH1 domain of the regulatory subunit 3 of Drosophila PP4, Falafel (Flfl), directly interacts with the centromeric protein C (CENP-C). Unlike other EVH1 domains that interact with proline-rich ligands, the crystal structure of the Flfl amino-terminal EVH1 domain bound to a CENP-C peptide reveals a new target-recognition mode for the phosphatase subunit. We also show that binding of Flfl to CENP-C is required to bring PP4 activity to centromeres to maintain CENP-C and attached core kinetochore proteins at chromosomes during mitosis.
- Department of Genetics, University of Cambridge, Downing Street, Cambridge CB2 3EH, UK.
Organizational Affiliation: