Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
Liu, W.D., Guo, R.T., Cui, Y.F., Chen, X., Wu, Q.Q., Zhu, D.M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N-acetylneuraminate lyase | 302 | Clostridioides difficile | Mutation(s): 0 Gene Names: nanA, CdNal EC: 4.1.3.3 | 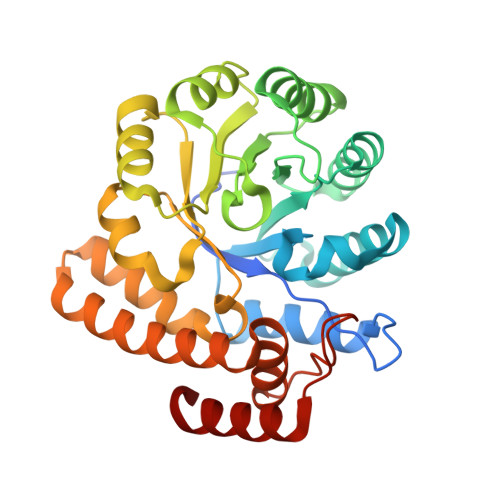 | |
UniProt | |||||
Find proteins for D5Q364 (Clostridioides difficile NAP08) Explore D5Q364 Go to UniProtKB: D5Q364 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D5Q364 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MN9 Query on MN9 | I [auth B], J [auth F] | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE C8 H15 N O6 MBLBDJOUHNCFQT-WCTZXXKLSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 81.476 | α = 90 |
| b = 121.604 | β = 96.12 |
| c = 121.065 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| HKL-2000 | data reduction |
| MOLREP | phasing |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China | China | 21072151 |
| National Natural Science Foundation of China | China | 21102100 |
| National Basic Research Program of China | China | 2011CB710801 |