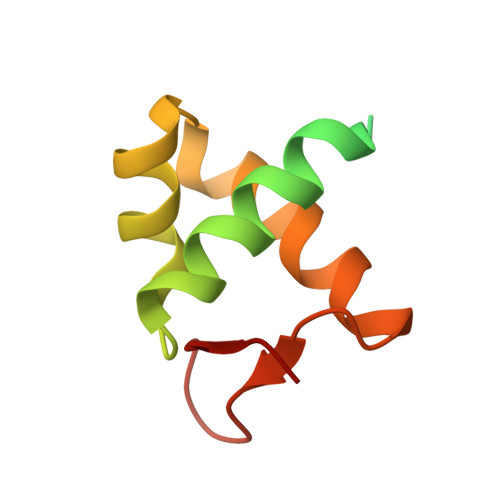
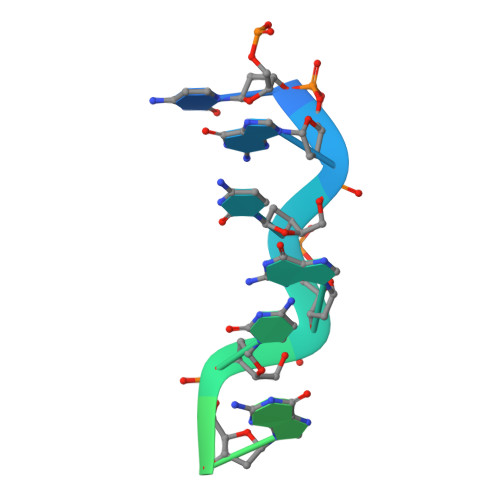
The Structure of the Cyprinid herpesvirus 3 ORF112-Z alpha Z-DNA Complex Reveals a Mechanism of Nucleic Acids Recognition Conserved with E3L, a Poxvirus Inhibitor of Interferon Response.
Kus, K., Rakus, K., Boutier, M., Tsigkri, T., Gabriel, L., Vanderplasschen, A., Athanasiadis, A.(2015) J Biological Chem 290: 30713-30725
- PubMed: 26559969
- DOI: https://doi.org/10.1074/jbc.M115.679407
- Primary Citation of Related Structures:
4WCG - PubMed Abstract:
In vertebrate species, the innate immune system down-regulates protein translation in response to viral infection through the action of the double-stranded RNA (dsRNA)-activated protein kinase (PKR). In some teleost species another protein kinase, Z-DNA-dependent protein kinase (PKZ), plays a similar role but instead of dsRNA binding domains, PKZ has Zα domains. These domains recognize the left-handed conformer of dsDNA and dsRNA known as Z-DNA/Z-RNA. Cyprinid herpesvirus 3 infects common and koi carp, which have PKZ, and encodes the ORF112 protein that itself bears a Zα domain, a putative competitive inhibitor of PKZ. Here we present the crystal structure of ORF112-Zα in complex with an 18-bp CpG DNA repeat, at 1.5 Å. We demonstrate that the bound DNA is in the left-handed conformation and identify key interactions for the specificity of ORF112. Localization of ORF112 protein in stress granules induced in Cyprinid herpesvirus 3-infected fish cells suggests a functional behavior similar to that of Zα domains of the interferon-regulated, nucleic acid surveillance proteins ADAR1 and DAI.
- From the Instituto Gulbenkian de Ciência, 2781-156, Oeiras, Portugal and.
Organizational Affiliation: