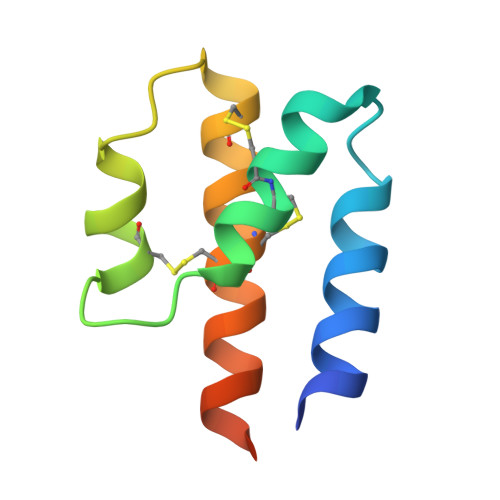
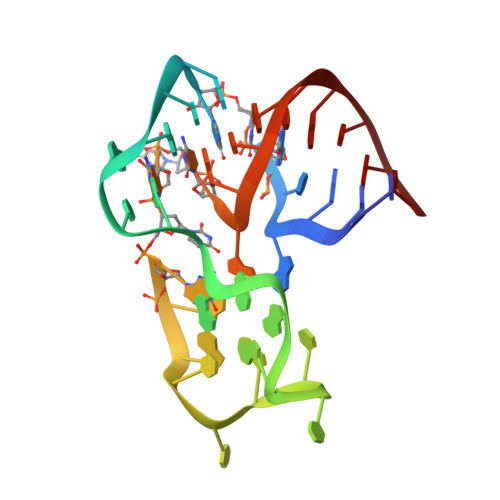
Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer.
Yatime, L., Maasch, C., Hoehlig, K., Klussmann, S., Andersen, G.R., Vater, A.(2015) Nat Commun 6: 6481-6481
- PubMed: 25901944
- DOI: https://doi.org/10.1038/ncomms7481
- Primary Citation of Related Structures:
4WB2, 4WB3 - PubMed Abstract:
L-Oligonucleotide aptamers (Spiegelmers) consist of non-natural L-configured nucleotides and are of particular therapeutic interest due to their high resistance to plasma nucleases. The anaphylatoxin C5a, a potent inflammatory mediator generated during complement activation that has been implicated with organ damage, can be efficiently targeted by Spiegelmers. Here, we present the first crystallographic structures of an active Spiegelmer, NOX-D20, bound to its physiological targets, mouse C5a and C5a-desArg. The structures reveal a complex 3D architecture for the L-aptamer that wraps around C5a, including an intramolecular G-quadruplex stabilized by a central Ca(2+) ion. Functional validation of the observed L-aptamer:C5a binding mode through mutational studies also rationalizes the specificity of NOX-D20 for mouse and human C5a against macaque and rat C5a. Finally, our structural model provides the molecular basis for the Spiegelmer affinity improvement through positional L-ribonucleotide to L-deoxyribonucleotide exchanges and for its inhibition of the C5a:C5aR interaction.
- Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 10C, DK-8000 Aarhus, Denmark.
Organizational Affiliation: