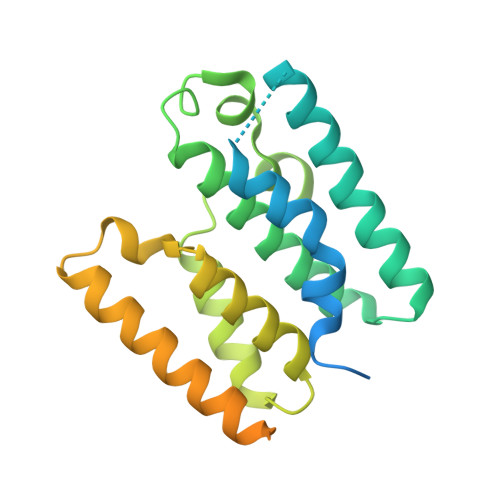
Crystal structure of the bacterial type VI secretion system component TssL from Vibrio cholerae.
Chang, J.H., Kim, Y.G.(2015) J Microbiol 53: 32-37
- PubMed: 25471186
- DOI: https://doi.org/10.1007/s12275-015-4539-0
- Primary Citation of Related Structures:
4V3I - PubMed Abstract:
The type VI secretion system (T6SS), commonly found in gram-negative bacteria, is responsible for exporting effector proteins. The T6SS has been reported to be cytotoxic to host cells. While the components and assembly of the T6SS complex have been largely assessed, structural data on T6SS components from virulent bacteria is remarkably insufficient. Here, we report the crystal structure of Vibrio cholerae TssL (VcTssL), a core component of T6SS. In spite of a relatively low sequence identity, the overall structure of VcTssL is largely similar to those from other bacterial homologs except for several differences found in local structural elements. A unique feature attributed to the C-terminal fragment of VcTssL is a crystallographic artifact. This incidental feature of VcTssL may provide insights into screening of molecular partners for the cytoplasmic domain of TssL. Additionally, our results may help in the design of molecular probes for a detailed understanding of the functional relationship between TssL and other T6SS components.
- Department of Biology, Teachers College, Kyungpook National University, Daegu, 702-701, Republic of Korea.
Organizational Affiliation: