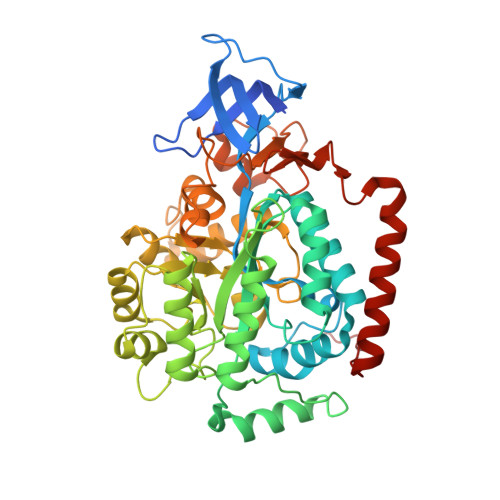
The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Peat, T.S., Newman, J., Balotra, S., Lucent, D., Warden, A.C., Scott, C.(2015) Acta Crystallogr D Biol Crystallogr 71: 710
- PubMed: 25760618
- DOI: https://doi.org/10.1107/S1399004715000619
- Primary Citation of Related Structures:
4V1X, 4V1Y - PubMed Abstract:
Atrazine chlorohydrolase (AtzA) was discovered and purified in the early 1990s from soil that had been exposed to the widely used herbicide atrazine. It was subsequently found that this enzyme catalyzes the first and necessary step in the breakdown of atrazine by the soil organism Pseudomonas sp. strain ADP. Although it has taken 20 years, a crystal structure of the full hexameric form of AtzA has now been obtained. AtzA is less well adapted to its physiological role (i.e. atrazine dechlorination) than the alternative metal-dependent atrazine chlorohydrolase (TrzN), with a substrate-binding pocket that is under considerable strain and for which the substrate is a poor fit.
- CSIRO Biomedical Manufacturing, Parkville, Australia.
Organizational Affiliation: