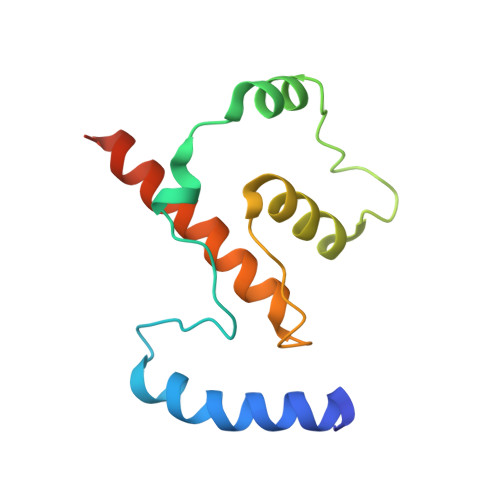
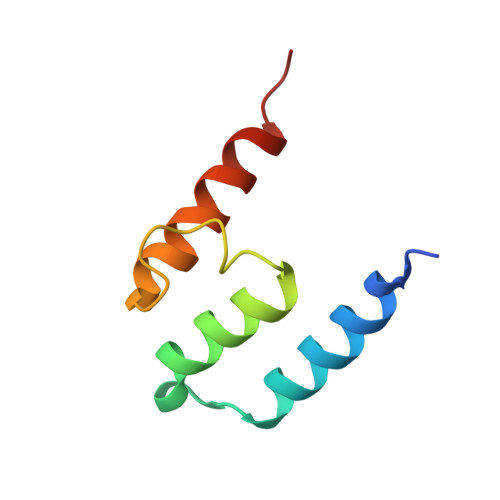
Crystal Structures of Ift70/52 and Ift52/46 Provide Insight Into Intraflagellar Transport B Core Complex Assembly.
Taschner, M., Kotsis, F., Braeuer, P., Kuehn, E.W., Lorentzen, E.(2014) J Cell Biol 207: 269
- PubMed: 25349261
- DOI: https://doi.org/10.1083/jcb.201408002
- Primary Citation of Related Structures:
4UZY, 4UZZ - PubMed Abstract:
Cilia are microtubule-based organelles that assemble via intraflagellar transport (IFT) and function as signaling hubs on eukaryotic cells. IFT relies on molecular motors and IFT complexes that mediate the contacts with ciliary cargo. To elucidate the architecture of the IFT-B complex, we reconstituted and purified the nonameric IFT-B core from Chlamydomonas reinhardtii and determined the crystal structures of C. reinhardtii IFT70/52 and Tetrahymena IFT52/46 subcomplexes. The 2.5-Å resolution IFT70/52 structure shows that IFT52330-370 is buried deeply within the IFT70 tetratricopeptide repeat superhelix. Furthermore, the polycystic kidney disease protein IFT88 binds IFT52281-329 in a complex that interacts directly with IFT70/IFT52330-381 in trans. The structure of IFT52C/IFT46C was solved at 2.3 Å resolution, and we show that it is essential for IFT-B core integrity by mediating interaction between IFT88/70/52/46 and IFT81/74/27/25/22 subcomplexes. Consistent with this, overexpression of mammalian IFT52C in MDCK cells is dominant-negative and causes IFT protein mislocalization and disrupted ciliogenesis. These data further rationalize several ciliogenesis phenotypes of IFT mutant strains.
- Department of Structural Cell Biology, Max-Planck-Institute of Biochemistry, D-82152 Martinsried, Germany.
Organizational Affiliation: