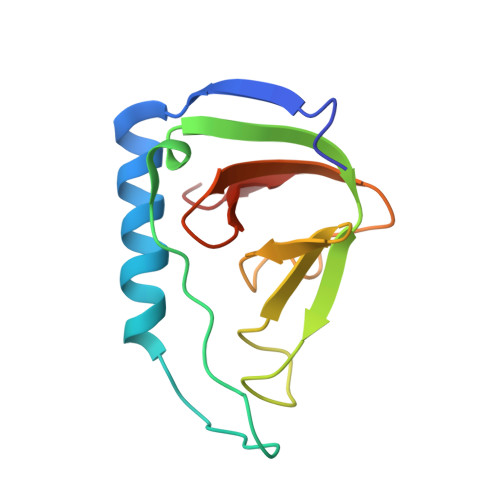
Solution Structure of a Soluble Fragment Derived from a Membrane Protein by Shotgun Proteolysis.
Allen, M.D., Christie, M., Jones, P., Porebski, B.T., Roome, B., Freund, S.M., Buckle, A.M., Bycroft, M., Christ, D.(2015) Protein Eng Des Sel 28: 445
- PubMed: 25877662
- DOI: https://doi.org/10.1093/protein/gzv021
- Primary Citation of Related Structures:
4UZM - PubMed Abstract:
We have previously reported a phage display method for the identification of protein domains on a genome-wide scale (shotgun proteolysis). Here we present the solution structure of a fragment of the Escherichia coli membrane protein yrfF, as identified by shotgun proteolysis, and determined by NMR spectroscopy. Despite the absence of computational predictions, the fragment formed a well-defined beta-barrel structure, distantly falling within the OB-fold classification. Our results highlight the potential of high-throughput experimental approaches for the identification of protein domains for structural studies.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK.
Organizational Affiliation: