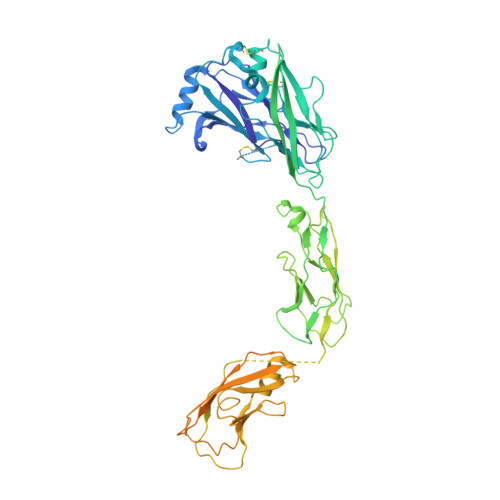
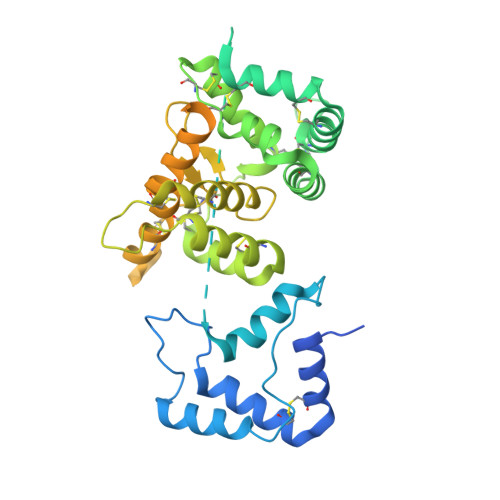
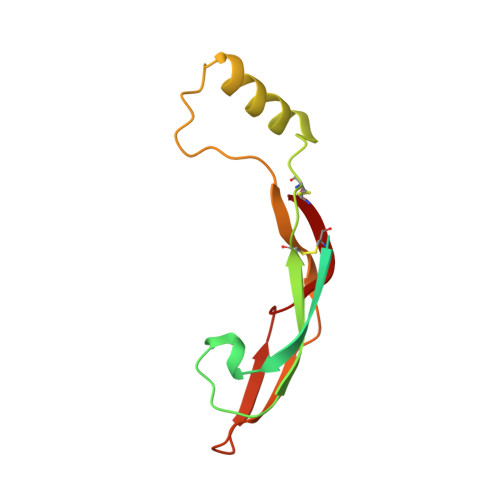
Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Goodman, K.M., Kjaer, S., Beuron, F., Knowles, P.P., Nawrotek, A., Burns, E.M., Purkiss, A.G., George, R., Santoro, M., Morris, E.P., Mcdonald, N.Q.(2014) Cell Rep 8: 1894
- PubMed: 25242331
- DOI: https://doi.org/10.1016/j.celrep.2014.08.040
- Primary Citation of Related Structures:
4UX8 - PubMed Abstract:
The RET receptor tyrosine kinase is essential to vertebrate development and implicated in multiple human diseases. RET binds a cell surface bipartite ligand comprising a GDNF family ligand and a GFRα coreceptor, resulting in RET transmembrane signaling. We present a hybrid structural model, derived from electron microscopy (EM) and low-angle X-ray scattering (SAXS) data, of the RET extracellular domain (RET(ECD)), GDNF, and GFRα1 ternary complex, defining the basis for ligand recognition. RET(ECD) envelopes the dimeric ligand complex through a composite binding site comprising four discrete contact sites. The GFRα1-mediated contacts are crucial, particularly close to the invariant RET calcium-binding site, whereas few direct contacts are made by GDNF, explaining how distinct ligand/coreceptor pairs are accommodated. The RET(ECD) cysteine-rich domain (CRD) contacts both ligand components and makes homotypic membrane-proximal interactions occluding three different antibody epitopes. Coupling of these CRD-mediated interactions suggests models for ligand-induced RET activation and ligand-independent oncogenic deregulation.
- Structural Biology Laboratory, Cancer Research UK, London Research Institute, 44 Lincoln's Inn Fields, London WC2A 3LY, UK.
Organizational Affiliation: