From Crystal Structure to in Silico Epitope Discovery in Burkholderia Pseudomallei Flagellar Hook-Associated Protein Flgk.
Gourlay, L.J., Thomas, R.J., Peri, C., Conchillo-Sole, O., Ferrer-Navarro, M., Nithichanon, A., Vila, J., Daura, X., Lertmemongkolchai, G., Titball, R., Colombo, G., Bolognesi, M.(2015) FEBS J 282: 1319
- PubMed: 25645451
- DOI: https://doi.org/10.1111/febs.13223
- Primary Citation of Related Structures:
4UT1 - PubMed Abstract:
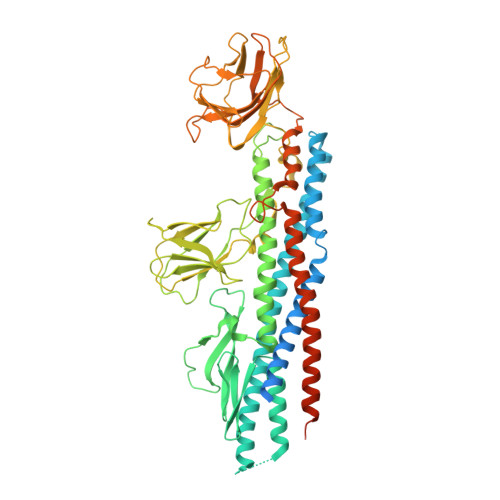
Melioidosis, caused by the Gram-negative bacterium Burkholderia pseudomallei, is a potentially fatal infection that is endemic in Southeast Asia and Northern Australia that is poorly controlled by antibiotics. Research efforts to identify antigenic components for a melioidosis vaccine have led to the identification of several proteins, including subunits forming the flagella that mediate bacterial motility, host colonization, and virulence. This study focuses on the B. pseudomallei flagellar hook-associated protein (FlgK(Bp)), and provides the first insights into the 3D structure of FlgK proteins as targets for structure-based antigen engineering. The FlgK(Bp) crystal structure (presented here at 1.8-Å resolution) reveals a multidomain fold, comprising two small β-domains protruding from a large elongated α-helical bundle core. The evident structural similarity to flagellin, the flagellar filament subunit protein, suggests that, depending on the bacterial species, flagellar hook-associated proteins are likely to show a conserved, elongated α-helical bundle scaffold coupled to a variable number of smaller domains. Furthermore, we present immune serum recognition data confirming, in agreement with previous findings, that recovered melioidosis patients produce elevated levels of antibodies against FlgK(Bp), in comparison with seronegative and seropositive healthy subjects. Moreover, we show that FlgK(Bp) has cytotoxic effects on cultured murine macrophages, suggesting an important role in bacterial pathogenesis. Finally, computational epitope prediction methods applied to the FlgK(Bp) crystal structure, coupled with in vitro mapping, allowed us to predict three antigenic regions that locate to discrete protein domains. Taken together, our results point to FlgK(Bp) as a candidate for the design and production of epitope-containing subunits/domains as potential vaccine components.
- Department of Biosciences, University of Milan, Italy.
Organizational Affiliation: