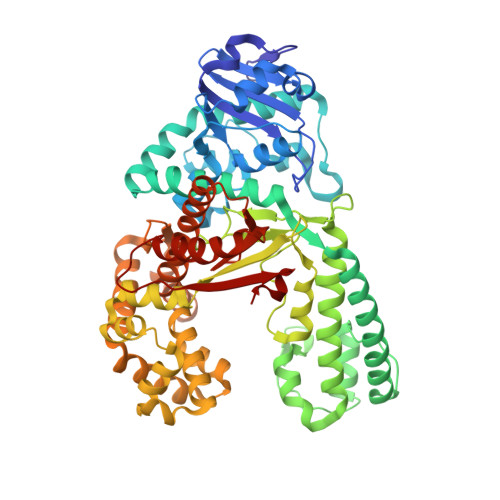
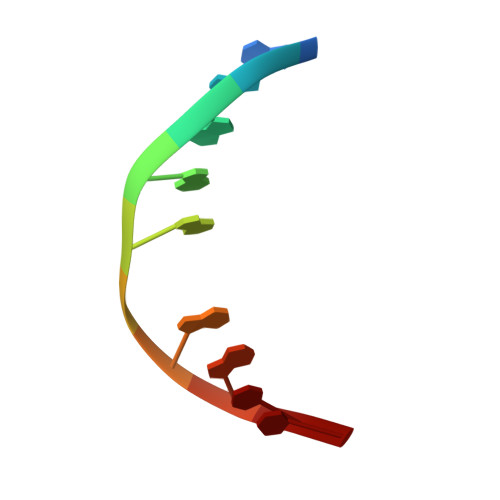
"Post-it" type connected DNA created with a reversible covalent cross-link.
Tomas-Gamasa, M., Serdjukow, S., Su, M., Muller, M., Carell, T.(2015) Angew Chem Int Ed Engl 54: 796-800
- PubMed: 25446281
- DOI: https://doi.org/10.1002/anie.201407854
- Primary Citation of Related Structures:
4UQG - PubMed Abstract:
We report the development of a new heterobase that is held together through reversible bonding. The so-formed cross-link adds strong stabilization to the DNA duplex. Despite this, the cross-link opens and closes through reversible imine bonding. Moreover, even enzymatic incorporation of the cross-link is possible. The new principle can be used to stabilize DNA duplexes and nanostructures that otherwise require high salt concentrations, which may hinder biological applications.
- Department of Chemistry and Pharmacy, Ludwig-Maximilians-Universität München, Butenandtstrasse 5-13, 81377 München (Germany) http://www.carellgroup.de.
Organizational Affiliation: