Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Anders, C., Niewoehner, O., Duerst, A., Jinek, M.(2014) Nature 513: 569
- PubMed: 25079318
- DOI: https://doi.org/10.1038/nature13579
- Primary Citation of Related Structures:
4UN3, 4UN4, 4UN5 - PubMed Abstract:
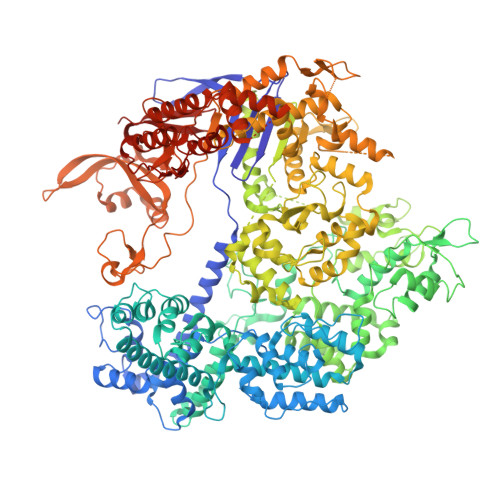
The CRISPR-associated protein Cas9 is an RNA-guided endonuclease that cleaves double-stranded DNA bearing sequences complementary to a 20-nucleotide segment in the guide RNA. Cas9 has emerged as a versatile molecular tool for genome editing and gene expression control. RNA-guided DNA recognition and cleavage strictly require the presence of a protospacer adjacent motif (PAM) in the target DNA. Here we report a crystal structure of Streptococcus pyogenes Cas9 in complex with a single-molecule guide RNA and a target DNA containing a canonical 5'-NGG-3' PAM. The structure reveals that the PAM motif resides in a base-paired DNA duplex. The non-complementary strand GG dinucleotide is read out via major-groove interactions with conserved arginine residues from the carboxy-terminal domain of Cas9. Interactions with the minor groove of the PAM duplex and the phosphodiester group at the +1 position in the target DNA strand contribute to local strand separation immediately upstream of the PAM. These observations suggest a mechanism for PAM-dependent target DNA melting and RNA-DNA hybrid formation. Furthermore, this study establishes a framework for the rational engineering of Cas9 enzymes with novel PAM specificities.
- Department of Biochemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland.
Organizational Affiliation: