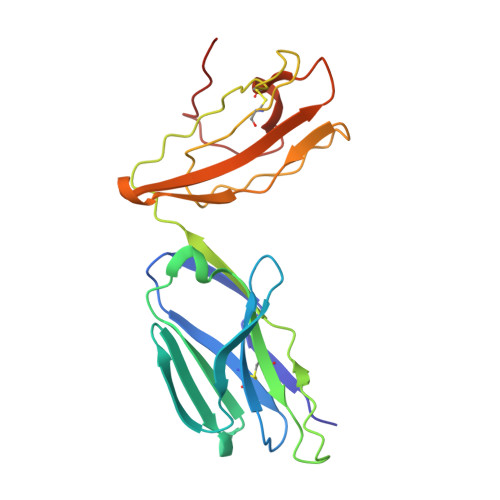
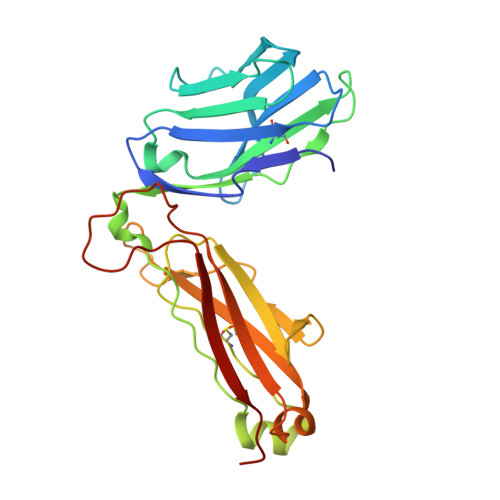
Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Rodstrom, K.E.J., Regenthal, P., Lindkvist-Petersson, K.(2015) PLoS One 10: 01318
- PubMed: 26147596
- DOI: https://doi.org/10.1371/journal.pone.0131988
- Primary Citation of Related Structures:
4UDT, 4UDU - PubMed Abstract:
T cells are crucial players in cell-mediated immunity. The specificity of their receptor, the T cell receptor (TCR), is central for the immune system to distinguish foreign from host antigens. Superantigens are bacterial toxins capable of inducing a toxic immune response by cross-linking the TCR and the major histocompatibility complex (MHC) class II and circumventing the antigen specificity. Here, we present the structure of staphylococcal enterotoxin E (SEE) in complex with a human T cell receptor, as well as the unligated T cell receptor structure. There are clear structural changes in the TCR loops upon superantigen binding. In particular, the HV4 loop moves to circumvent steric clashes upon complex formation. In addition, a predicted ternary model of SEE in complex with both TCR and MHC class II displays intermolecular contacts between the TCR α-chain and the MHC, suggesting that the TCR α-chain is of importance for complex formation.
- Department of Experimental Medical Science, Lund University, BMC C13, 22 184, Lund, Sweden.
Organizational Affiliation: