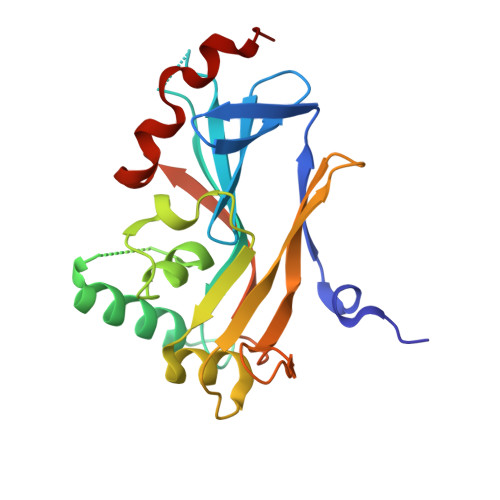
The Unusual Fold of Herpes Simplex Virus 1 UL21, a Multifunctional Tegument Protein.
Metrick, C.M., Chadha, P., Heldwein, E.E.(2015) J Virol 89: 2979-2984
- PubMed: 25540382
- DOI: https://doi.org/10.1128/JVI.03516-14
- Primary Citation of Related Structures:
4U4H - PubMed Abstract:
UL21 is a conserved protein in the tegument of alphaherpesviruses and has multiple important albeit poorly understood functions in viral replication and pathogenesis. To provide a roadmap for exploration of the multiple roles of UL21, we determined the crystal structure of its conserved N-terminal domain from herpes simplex virus 1 to 2.0-Å resolution, which revealed a novel sail-like protein fold. Evolutionarily conserved surface patches highlight residues of potential importance for future targeting by mutagenesis.
- Department of Molecular Biology and Microbiology and Graduate Program in Biochemistry, Sackler School of Graduate Biomedical Sciences, Tufts University School of Medicine, Boston, Massachusetts, USA.
Organizational Affiliation: