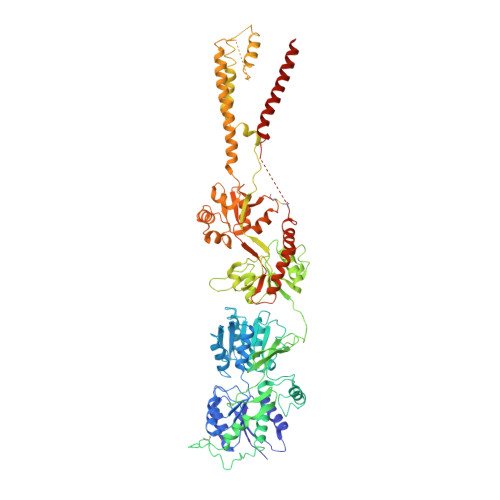
Structure of an agonist-bound ionotropic glutamate receptor.
Yelshanskaya, M.V., Li, M., Sobolevsky, A.I.(2014) Science 345: 1070-1074
- PubMed: 25103407
- DOI: https://doi.org/10.1126/science.1256508
- Primary Citation of Related Structures:
4U4F, 4U4G - PubMed Abstract:
Ionotropic glutamate receptors (iGluRs) mediate most excitatory neurotransmission in the central nervous system and function by opening their ion channel in response to binding of agonist glutamate. Here, we report a structure of a homotetrameric rat GluA2 receptor in complex with partial agonist (S)-5-nitrowillardiine. Comparison of this structure with the closed-state structure in complex with competitive antagonist ZK 200775 suggests conformational changes that occur during iGluR gating. Guided by the structures, we engineered disulfide cross-links to probe domain interactions that are important for iGluR gating events. The combination of structural information, kinetic modeling, and biochemical and electrophysiological experiments provides insight into the mechanism of iGluR gating.
- Department of Biochemistry and Molecular Biophysics, Columbia University, 650 West 168th Street, New York, NY 10032, USA.
Organizational Affiliation: