Crystal structure of Coh3ScaB-XDoc_M1ScaA complex: A N-terminal interface mutant of type II Cohesin-X-Dockerin complex from Acetivibrio cellulolyticus
Alves, V.D., Cameron, K., Najmudin, S.H., Fontes, C.M.G.A.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cellulosomal scaffoldin adaptor protein B | 168 | Acetivibrio cellulolyticus | Mutation(s): 0 Gene Names: scaB | 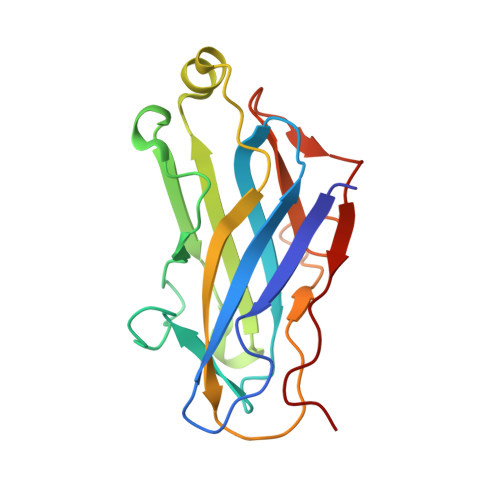 | |
UniProt | |||||
Find proteins for Q7WYN3 (Acetivibrio cellulolyticus) Explore Q7WYN3 Go to UniProtKB: Q7WYN3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7WYN3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cellulosomal scaffoldin | 184 | Acetivibrio cellulolyticus | Mutation(s): 1 Gene Names: cipV EC: 3.2.1.4 | 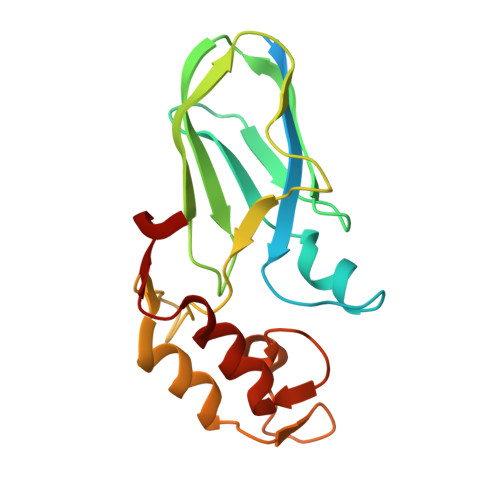 | |
UniProt | |||||
Find proteins for Q9RPL0 (Acetivibrio cellulolyticus) Explore Q9RPL0 Go to UniProtKB: Q9RPL0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9RPL0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NHE Query on NHE | C [auth A], D [auth A], G [auth B] | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID C8 H17 N O3 S MKWKNSIESPFAQN-UHFFFAOYSA-N |  | ||
| CA Query on CA | E [auth B], F [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 72.32 | α = 90 |
| b = 72.32 | β = 90 |
| c = 231.49 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Aimless | data scaling |
| BALBES | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| FCT | Portugal | EXPL/BIA-MIC/1176/2012 |