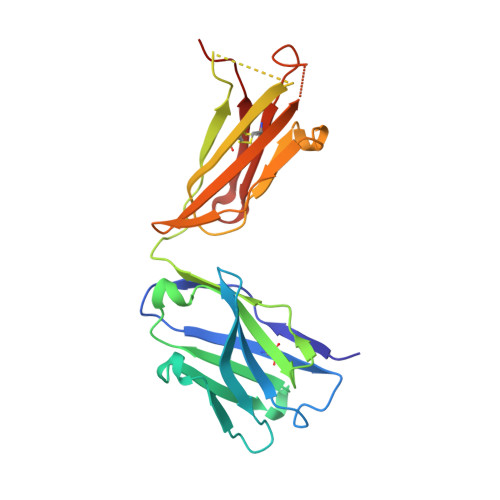
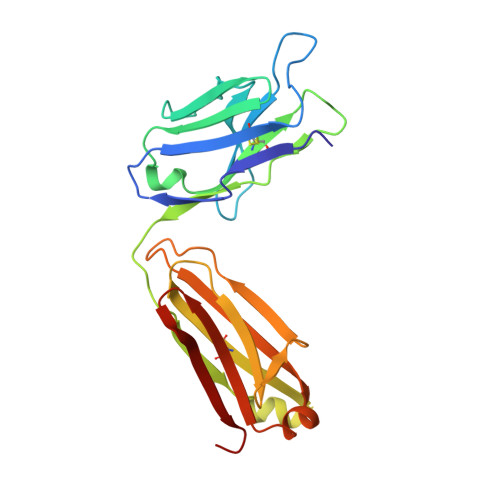
Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Horwacik, I., Golik, P., Grudnik, P., Kolinski, M., Zdzalik, M., Rokita, H., Dubin, G.(2015) Mol Cell Proteomics 14: 2577-2590
- PubMed: 26179345
- DOI: https://doi.org/10.1074/mcp.M115.052720
- Primary Citation of Related Structures:
4TRP, 4TUJ, 4TUK, 4TUL, 4TUO - PubMed Abstract:
Monoclonal antibodies targeting GD2 ganglioside (GD2) have recently been approved for the treatment of high risk neuroblastoma and are extensively evaluated in clinics in other indications. This study illustrates how a therapeutic antibody distinguishes between different types of gangliosides present on normal and cancer cells and informs how synthetic peptides can imitate ganglioside in its binding to the antibody. Using high resolution crystal structures we demonstrate that the ganglioside recognition by a model antibody (14G2a) is based primarily on an extended network of direct and water molecule mediated hydrogen bonds. Comparison of the GD2-Fab structure with that of a ligand free antibody reveals an induced fit mechanism of ligand binding. These conclusions are validated by directed mutagenesis and allowed structure guided generation of antibody variant with improved affinity toward GD2. Contrary to the carbohydrate, both evaluated mimetic peptides utilize a "key and lock" interaction mechanism complementing the surface of the antibody binding groove exactly as found in the empty structure. The interaction of both peptides with the Fab relies considerably on hydrophobic contacts however, the detailed connections differ significantly between the peptides. As such, the evaluated peptide carbohydrate mimicry is defined primarily in a functional and not in structural manner.
- From the ‡Laboratory of Molecular Genetics and Virology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, 7 Gronostajowa St., 30-387 Krakow, Poland; irena.horwacik@uj.edu.pl.
Organizational Affiliation: