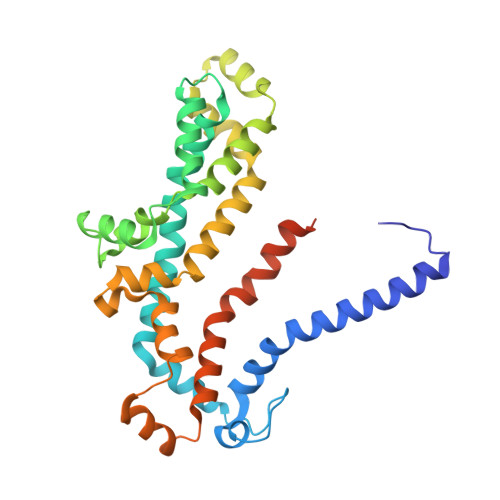
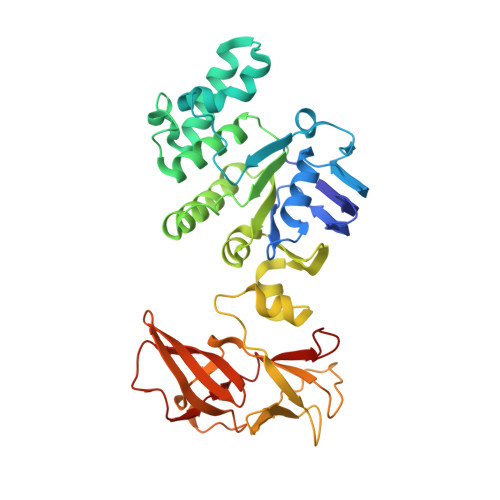
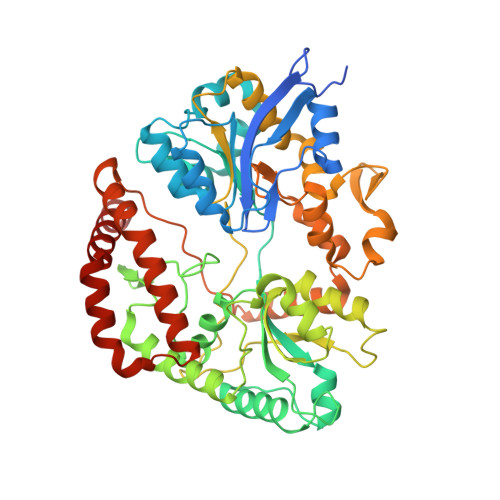
Structure of a Bacterial ABC Transporter Involved in the Import of an Acidic Polysaccharide Alginate
Maruyama, Y., Itoh, T., Kaneko, A., Nishitani, Y., Mikami, B., Hashimoto, W., Murata, K.(2015) Structure 23: 1643-1654
- PubMed: 26235029
- DOI: https://doi.org/10.1016/j.str.2015.06.021
- Primary Citation of Related Structures:
4TQU, 4TQV - PubMed Abstract:
The acidic polysaccharide alginate represents a promising marine biomass for the microbial production of biofuels, although the molecular and structural characteristics of alginate transporters remain to be clarified. In Sphingomonas sp. A1, the ATP-binding cassette transporter AlgM1M2SS is responsible for the import of alginate across the cytoplasmic membrane. Here, we present the substrate-transport characteristics and quaternary structure of AlgM1M2SS. The addition of poly- or oligoalginate enhanced the ATPase activity of reconstituted AlgM1M2SS coupled with one of the periplasmic solute-binding proteins, AlgQ1 or AlgQ2. External fluorescence-labeled oligoalginates were specifically imported into AlgM1M2SS-containing proteoliposomes in the presence of AlgQ2, ATP, and Mg(2+). The crystal structure of AlgQ2-bound AlgM1M2SS adopts an inward-facing conformation. The interaction between AlgQ2 and AlgM1M2SS induces the formation of an alginate-binding tunnel-like structure accessible to the solvent. The translocation route inside the transmembrane domains contains charged residues suitable for the import of acidic saccharides.
- Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan.
Organizational Affiliation: