X-ray Transparent Microfluidic Chip for Mesophase-Based Crystallization of Membrane Proteins and On-Chip Structure Determination.
Khvostichenko, D.S., Schieferstein, J.M., Pawate, A.S., Laible, P.D., Kenis, P.J.(2014) Cryst Growth Des 14: 4886-4890
- PubMed: 25285049
- DOI: https://doi.org/10.1021/cg5011488
- Primary Citation of Related Structures:
4TQQ - PubMed Abstract:
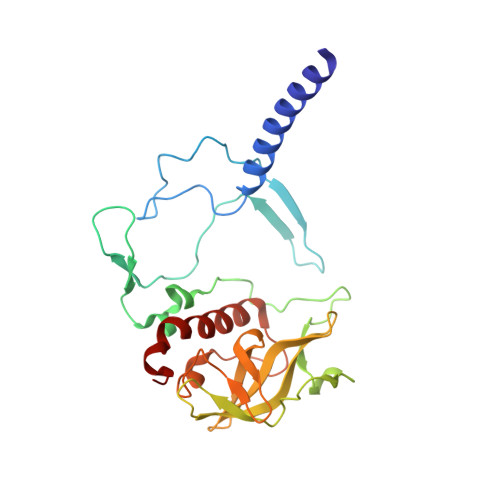
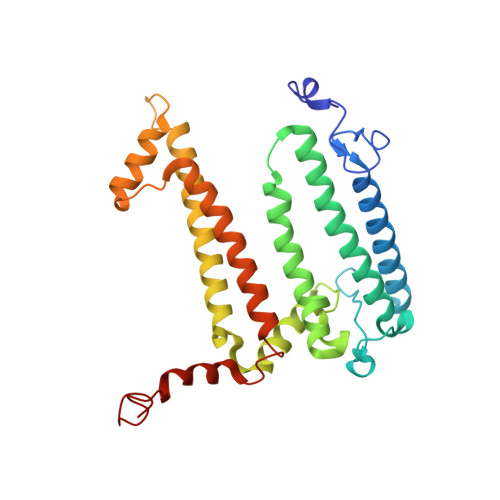
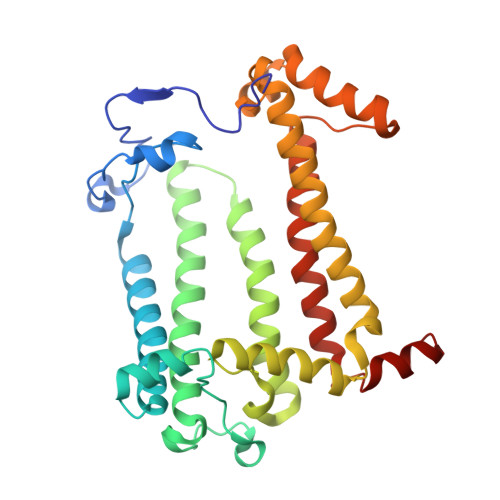
Crystallization from lipidic mesophase matrices is a promising route to diffraction-quality crystals and structures of membrane proteins. The microfluidic approach reported here eliminates two bottlenecks of the standard mesophase-based crystallization protocols: (i) manual preparation of viscous mesophases and (ii) manual harvesting of often small and fragile protein crystals. In the approach reported here, protein-loaded mesophases are formulated in an X-ray transparent microfluidic chip using only 60 nL of the protein solution per crystallization trial. The X-ray transparency of the chip enables diffraction data collection from multiple crystals residing in microfluidic wells, eliminating the normally required manual harvesting and mounting of individual crystals. We validated our approach by on-chip crystallization of photosynthetic reaction center, a membrane protein from Rhodobacter sphaeroides , followed by solving its structure to a resolution of 2.5 Å using X-ray diffraction data collected on-chip under ambient conditions. A moderate conformational change in hydrophilic chains of the protein was observed when comparing the on-chip, room temperature structure with known structures for which data were acquired under cryogenic conditions.
- Department of Chemical & Biomolecular Engineering, University of Illinois at Urbana-Champaign , 600 South Mathews Avenue, Urbana, Illinois 61801, United States.
Organizational Affiliation: