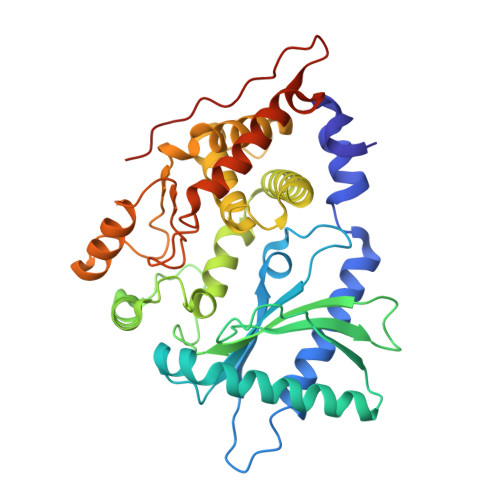
The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Lohofener, J., Steinke, N., Kay-Fedorov, P., Baruch, P., Nikulin, A., Tishchenko, S., Manstein, D.J., Fedorov, R.(2015) Structure 23: 851-862
- PubMed: 25892109
- DOI: https://doi.org/10.1016/j.str.2015.03.012
- Primary Citation of Related Structures:
4RWN, 4RWO, 4RWP, 4RWQ - PubMed Abstract:
2'-5'-Oligoadenylate synthetases (OASs) produce the second messenger 2'-5'-oligoadenylate, which activates RNase L to induce an intrinsic antiviral state. We report on the crystal structures of catalytic intermediates of OAS1 including the OAS1·dsRNA complex without substrates, with a donor substrate, and with both donor and acceptor substrates. Combined with kinetic studies of point mutants and the previously published structure of the apo form of OAS1, the new data suggest a sequential mechanism of OAS activation and show the individual roles of each component. They reveal a dsRNA-mediated push-pull effect responsible for large conformational changes in OAS1, the catalytic role of the active site Mg(2+), and the structural basis for the 2'-specificity of product formation. Our data reveal similarities and differences in the activation mechanisms of members of the OAS/cyclic GMP-AMP synthase family of innate immune sensors. In particular, they show how helix 3103-α5 blocks the synthesis of cyclic dinucleotides by OAS1.
- Institute for Biophysical Chemistry, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany.
Organizational Affiliation: