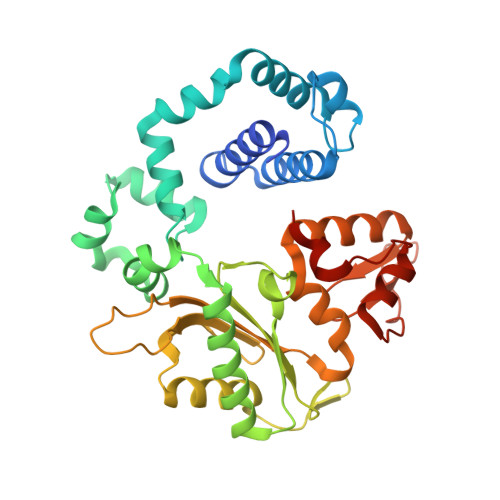
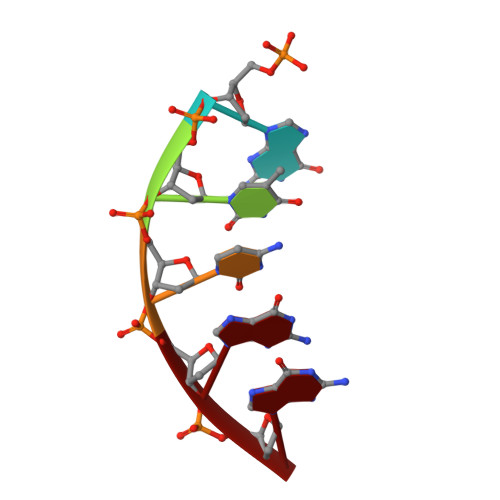
Viewing Human DNA Polymerase beta Faithfully and Unfaithfully Bypass an Oxidative Lesion by Time-Dependent Crystallography.
Vyas, R., Reed, A.J., Tokarsky, E.J., Suo, Z.(2015) J Am Chem Soc 137: 5225-5230
- PubMed: 25825995
- DOI: https://doi.org/10.1021/jacs.5b02109
- Primary Citation of Related Structures:
4RPX, 4RPY, 4RPZ, 4RQ0, 4RQ1, 4RQ2, 4RQ3, 4RQ4, 4RQ5, 4RQ6, 4RQ7, 4RQ8 - PubMed Abstract:
One common oxidative DNA lesion, 8-oxo-7,8-dihydro-2'-deoxyguanine (8-oxoG), is highly mutagenic in vivo due to its anti-conformation forming a Watson-Crick base pair with correct deoxycytidine 5'-triphosphate (dCTP) and its syn-conformation forming a Hoogsteen base pair with incorrect deoxyadenosine 5'-triphosphate (dATP). Here, we utilized time-resolved X-ray crystallography to follow 8-oxoG bypass by human DNA polymerase β (hPolβ). In the 12 solved structures, both Watson-Crick (anti-8-oxoG:anti-dCTP) and Hoogsteen (syn-8-oxoG:anti-dATP) base pairing were clearly visible and were maintained throughout the chemical reaction. Additionally, a third Mg(2+) appeared during the process of phosphodiester bond formation and was located between the reacting α- and β-phosphates of the dNTP, suggesting its role in stabilizing reaction intermediates. After phosphodiester bond formation, hPolβ reopened its conformation, pyrophosphate was released, and the newly incorporated primer 3'-terminal nucleotide stacked, rather than base paired, with 8-oxoG. These structures provide the first real-time pictures, to our knowledge, of how a polymerase correctly and incorrectly bypasses a DNA lesion.
- †Department of Chemistry and Biochemistry, ‡The Ohio State Biochemistry and §Biophysics Programs, The Ohio State University, Columbus, Ohio 43210, United States.
Organizational Affiliation: