The structure of Synechococcus elongatus enolase reveals key aspects of phosphoenolpyruvate binding
Gonzalez, J.M., Marti-Arbona, R., Chen, J.C.H., Unkefer, C.J.(2022) Acta Crystallogr F Struct Biol Commun 78: 177-184
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2022) Acta Crystallogr F Struct Biol Commun 78: 177-184
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Enolase | 424 | Synechococcus elongatus PCC 7942 = FACHB-805 | Mutation(s): 0 Gene Names: eno EC: 4.2.1.11 | 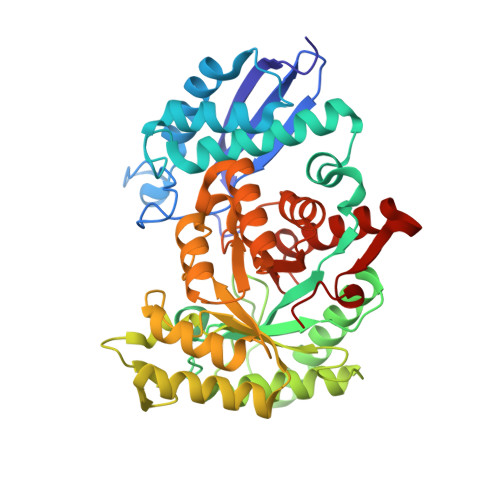 | |
UniProt | |||||
Find proteins for Q31QJ8 (Synechococcus elongatus (strain ATCC 33912 / PCC 7942 / FACHB-805)) Explore Q31QJ8 Go to UniProtKB: Q31QJ8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q31QJ8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CA Query on CA | B [auth A], C [auth A], D [auth A], E [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 164.282 | α = 90 |
| b = 164.282 | β = 90 |
| c = 75.5 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Scientific and Technical Research Council (CONICET) | Argentina | PIP0024 |