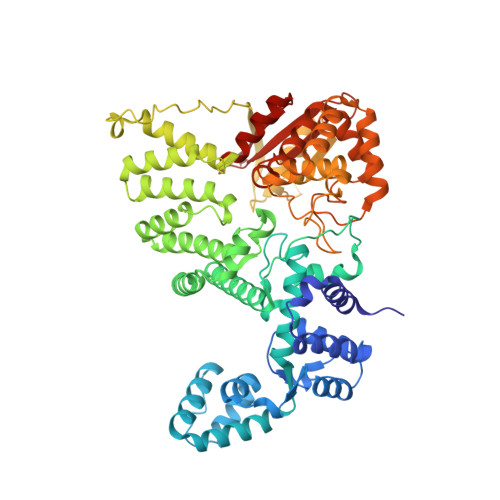
DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Wang, R., Persky, N.S., Yoo, B., Ouerfelli, O., Smogorzewska, A., Elledge, S.J., Pavletich, N.P.(2014) Science 346: 1127-1130
- PubMed: 25430771
- DOI: https://doi.org/10.1126/science.1258973
- Primary Citation of Related Structures:
4RI8, 4RI9, 4RIA, 4RIB, 4RIC, 4RID - PubMed Abstract:
DNA interstrand cross-links (ICLs) are highly toxic lesions associated with cancer and degenerative diseases. ICLs can be repaired by the Fanconi anemia (FA) pathway and through FA-independent processes involving the FAN1 nuclease. In this work, FAN1-DNA crystal structures and biochemical data reveal that human FAN1 cleaves DNA successively at every third nucleotide. In vitro, this exonuclease mechanism allows FAN1 to excise an ICL from one strand through flanking incisions. DNA access requires a 5'-terminal phosphate anchor at a nick or a 1- or 2-nucleotide flap and is augmented by a 3' flap, suggesting that FAN1 action is coupled to DNA synthesis or recombination. FAN1's mechanism of ICL excision is well suited for processing other localized DNA adducts as well.
- Structural Biology Program and Howard Hughes Medical Institute, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: