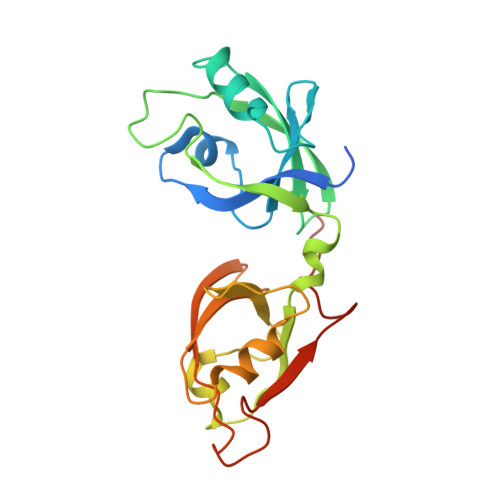
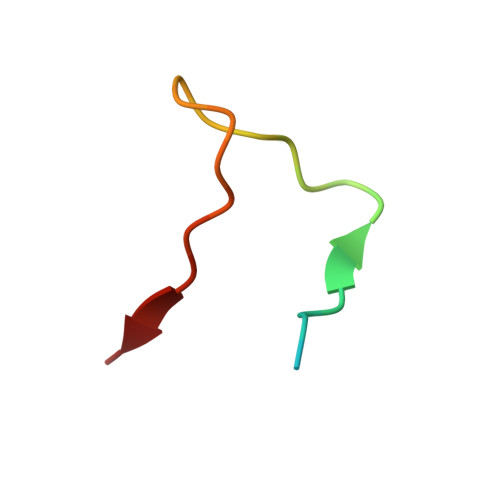
Structural Basis for the Interaction between the Golgi Reassembly-stacking Protein GRASP65 and the Golgi Matrix Protein GM130.
Hu, F., Shi, X., Li, B., Huang, X., Morelli, X., Shi, N.(2015) J Biological Chem 290: 26373-26382
- PubMed: 26363069
- DOI: https://doi.org/10.1074/jbc.M115.657940
- Primary Citation of Related Structures:
4REY - PubMed Abstract:
GM130 and GRASP65 are Golgi peripheral membrane proteins that play a key role in Golgi stacking and vesicle tethering. However, the molecular details of their interaction and their structural role as a functional unit remain unclear. Here, we present the crystal structure of the PDZ domains of GRASP65 in complex with the GM130 C-terminal peptide at 1.96-Å resolution. In contrast to previous findings proposing that GM130 interacts with GRASP65 at the PDZ2 domain only, our crystal structure of the complex indicates that GM130 binds to GRASP65 at two distinct sites concurrently and that both the PDZ1 and PDZ2 domains of GRASP65 participate in this molecular interaction. Mutagenesis experiments support these structural observations and demonstrate that they are required for GRASP65-GM130 association.
- From the State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou 350002, China and.
Organizational Affiliation: