Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Zhao, Q., Xue, X., Longerich, S., Sung, P., Xiong, Y.(2014) Nat Commun 5: 5726-5726
- PubMed: 25500724
- DOI: https://doi.org/10.1038/ncomms6726
- Primary Citation of Related Structures:
4REA, 4REB, 4REC - PubMed Abstract:
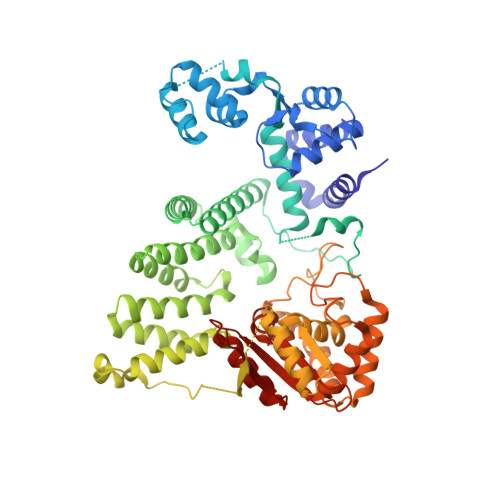
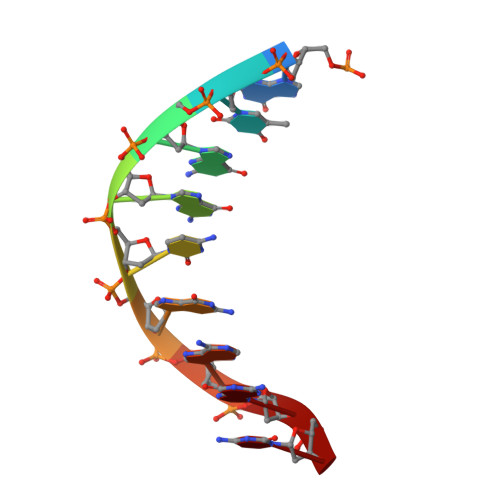
Human FANCD2-associated nuclease 1 (FAN1) is a DNA structure-specific nuclease involved in the processing of DNA interstrand crosslinks (ICLs). FAN1 maintains genomic stability and prevents tissue decline in multiple organs, yet it confers ICL-induced anti-cancer drug resistance in several cancer subtypes. Here we report three crystal structures of human FAN1 in complex with a 5' flap DNA substrate, showing that two FAN1 molecules form a head-to-tail dimer to locate the lesion, orient the DNA and unwind a 5' flap for subsequent incision. Biochemical experiments further validate our model for FAN1 action, as structure-informed mutations that disrupt protein dimerization, substrate orientation or flap unwinding impair the structure-specific nuclease activity. Our work elucidates essential aspects of FAN1-DNA lesion recognition and a unique mechanism of incision. These structural insights shed light on the cellular mechanisms underlying organ degeneration protection and cancer drug resistance mediated by FAN1.
- Department of Molecular Biophysics and Biochemistry, Yale University School of Medicine, New Haven, Connecticut 06520, USA.
Organizational Affiliation: