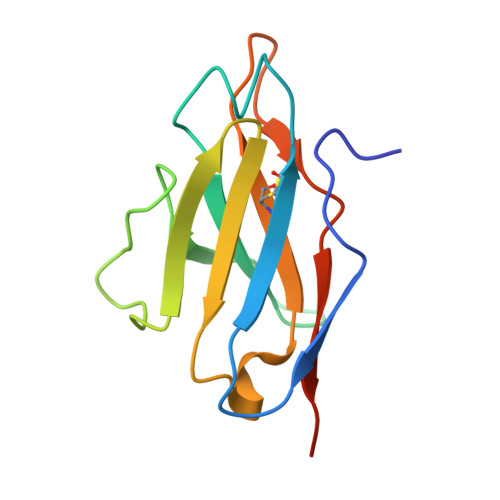
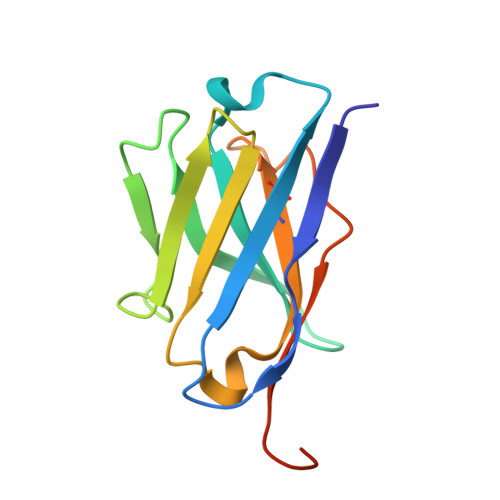
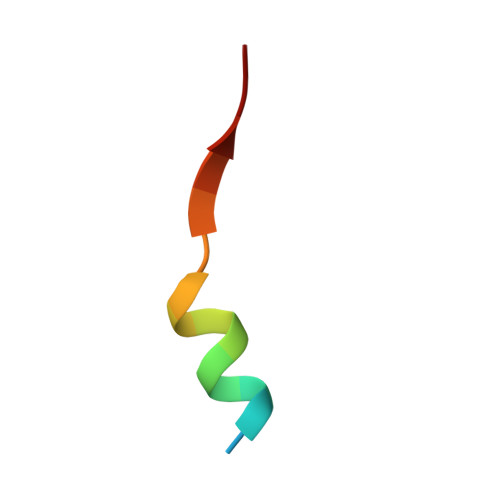Structure of a single-chain fv bound to the 17 N-terminal residues of huntingtin provides insights into pathogenic amyloid formation and suppression.
De Genst, E., Chirgadze, D.Y., Klein, F.A., Butler, D.C., Matak-Vinkovic, D., Trottier, Y., Huston, J.S., Messer, A., Dobson, C.M.(2015) J Mol Biology 427: 2166-2178
- PubMed: 25861763
- DOI: https://doi.org/10.1016/j.jmb.2015.03.021
- Primary Citation of Related Structures:
4RAV - PubMed Abstract:
Huntington's disease is triggered by misfolding of fragments of mutant forms of the huntingtin protein (mHTT) with aberrant polyglutamine expansions. The C4 single-chain Fv antibody (scFv) binds to the first 17 residues of huntingtin [HTT(1-17)] and generates substantial protection against multiple phenotypic pathologies in situ and in vivo. We show in this paper that C4 scFv inhibits amyloid formation by exon1 fragments of huntingtin in vitro and elucidate the structural basis for this inhibition and protection by determining the crystal structure of the complex of C4 scFv and HTT(1-17). The peptide binds with residues 3-11 forming an amphipathic helix that makes contact with the antibody fragment in such a way that the hydrophobic face of this helix is shielded from the solvent. Residues 12-17 of the peptide are in an extended conformation and interact with the same region of another C4 scFv:HTT(1-17) complex in the asymmetric unit, resulting in a β-sheet interface within a dimeric C4 scFv:HTT(1-17) complex. The nature of this scFv-peptide complex was further explored in solution by high-resolution NMR and physicochemical analysis of species in solution. The results provide insights into the manner in which C4 scFv inhibits the aggregation of HTT, and hence into its therapeutic potential, and suggests a structural basis for the initial interactions that underlie the formation of disease-associated amyloid fibrils by HTT.
- Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK. Electronic address: ejjd2@cam.ac.uk.
Organizational Affiliation: