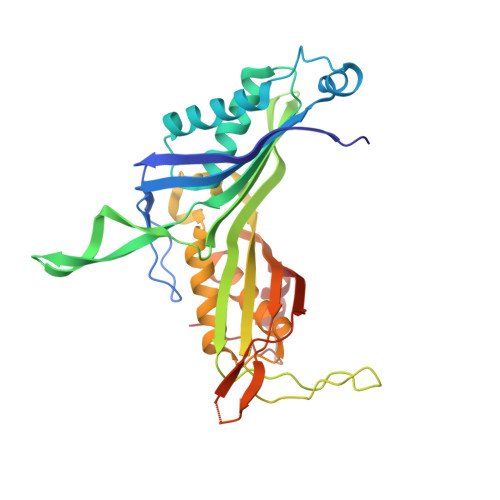
Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Feng, J., Wang, L., Liu, H., Yang, X., Liu, L., Xie, Y., Liu, M., Zhao, Y., Li, X., Wang, D., Zhan, C.G., Liao, F.(2015) Appl Microbiol Biotechnol 99: 7973-7986
- PubMed: 25786739
- DOI: https://doi.org/10.1007/s00253-015-6520-6
- Primary Citation of Related Structures:
4R8X, 4R99 - PubMed Abstract:
Bacillus fastidious uricase (BF uricase) containing 322 amino acid residues exhibited high stability under physiological conditions. Its crystal structure was solved to 1.4-Å resolution, showing homotetramer containing two homodimers. After the intersubunit antiparallel β-sheet in its homodimer, each subunit had a total of 18 C-terminus residues forming an α-helix (Q305-A313) and random coil (S314-L322) on surface to bury other two α-helices (I227-T238 and I244-R258). In comparison, reported crystal structures of Arthrobacter globiformis and Aspergillus flavus uricases had atomic coordinates of only some C-terminus residues, while the crystal structures of all the other uricases accessible before September 2014 missed atomic coordinates of all their C-terminus residues, after the intersubunit antiparallel β-sheets. In each homodimer of BF uricase, H-bonds were found between E311 and Y249 and between Y319 and D257; electrostatic interaction networks were found to surround D307 plus R310 and intersubunit R3, K312 plus D257, E318 plus K242, and L322 plus R258. Amino acid mutations that disrupted those interactions when R3 and D307 were reserved caused moderate decreases of activity at pH 9.2 while negligible decreases of activity at pH 7.4, but destroyed stability at pH 7.4 while slightly decreased stability at pH 9.2. Such structural information guided the fusion of 6His-tag to the C-terminus of the mutant L322D with SNSNSN as a linker to reserve the activity and stability. Hence, the folding of the C-terminus residues is crucial for thermal stability of BF uricase under physiological conditions; these new structural insights are valuable for molecular engineering of uricases.
- Key Laboratory of Medical Laboratory Diagnostics of the Education Ministry, College of Laboratory Medicine, Chongqing Medical University, Chongqing, 400016, China.
Organizational Affiliation: