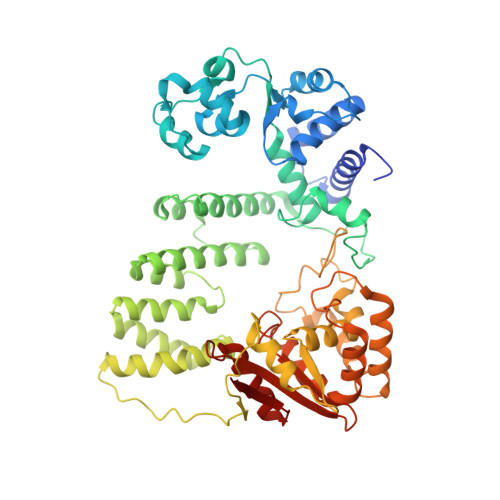
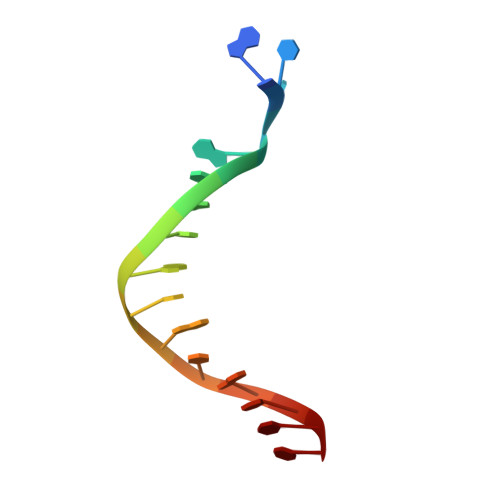
Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Gwon, G.H., Kim, Y.R., Liu, Y., Watson, A.T., Jo, A., Etheridge, T.J., Yuan, F., Zhang, Y., Kim, Y.C., Carr, A.M., Cho, Y.(2014) Genes Dev 28: 2276-2290
- PubMed: 25319828
- DOI: https://doi.org/10.1101/gad.248492.114
- Primary Citation of Related Structures:
4R89, 4R8A - PubMed Abstract:
Fanconi anemia (FA) is an autosomal recessive genetic disorder caused by defects in any of 15 FA genes responsible for processing DNA interstrand cross-links (ICLs). The ultimate outcome of the FA pathway is resolution of cross-links, which requires structure-selective nucleases. FA-associated nuclease 1 (FAN1) is believed to be recruited to lesions by a monoubiquitinated FANCI-FANCD2 (ID) complex and participates in ICL repair. Here, we determined the crystal structure of Pseudomonas aeruginosa FAN1 (PaFAN1) lacking the UBZ (ubiquitin-binding zinc) domain in complex with 5' flap DNA. All four domains of the right-hand-shaped PaFAN1 are involved in DNA recognition, with each domain playing a specific role in bending DNA at the nick. The six-helix bundle that binds the junction connects to the catalytic viral replication and repair (VRR) nuclease (VRR nuc) domain, enabling FAN1 to incise the scissile phosphate a few bases distant from the junction. The six-helix bundle also inhibits the cleavage of intact Holliday junctions. PaFAN1 shares several conserved features with other flap structure-selective nucleases despite structural differences. A clamping motion of the domains around the wedge helix, which acts as a pivot, facilitates nucleolytic cleavage. The PaFAN1 structure provides insights into how archaeal Holliday junction resolvases evolved to incise 5' flap substrates and how FAN1 integrates with the FA complex to participate in ICL repair.
- Department of Life Science, Pohang University of Science and Technology, Pohang 790-784, South Korea;
Organizational Affiliation: