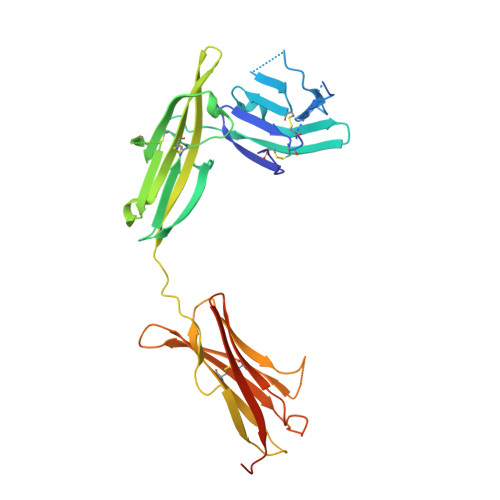
Structural basis for the specific recognition of IL-18 by its alpha receptor.
Wei, H., Wang, D., Qian, Y., Liu, X., Fan, S., Yin, H.S., Wang, X.(2014) FEBS Lett 588: 3838-3843
- PubMed: 25261253
- DOI: https://doi.org/10.1016/j.febslet.2014.09.019
- Primary Citation of Related Structures:
4R6U - PubMed Abstract:
Interleukin 18 (IL-18), a member of the IL-1 family of cytokines, is an important regulator of innate and acquired immune responses. It signals through its ligand-binding primary receptor IL-18Rα and accessory receptor IL-18Rβ. Here we report the crystal structure of IL-18 with the ectodomain of IL-18Rα, which reveals the structural basis for their specific recognition. It confirms that surface charge complementarity determines the ligand-binding specificity of primary receptors in the IL-1 receptor family. We suggest that IL-18 signaling complex adopts an architecture similar to other agonistic cytokines and propose a general ligand-receptor assembly and activation model for the IL-1 family.
- Ministry of Education Key Laboratory of Protein Science, Center for Structural Biology, Collaborative Innovation Center for Biotherapy, School of Life Sciences, Tsinghua University, Beijing 100084, China; Collaborative Innovation Center for Biotherapy, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, Sichuan University, Chengdu, China.
Organizational Affiliation: