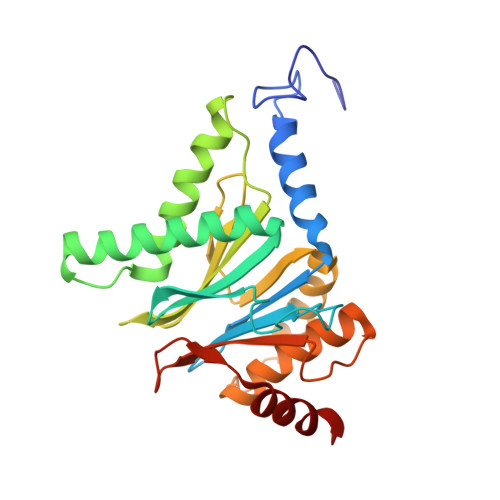
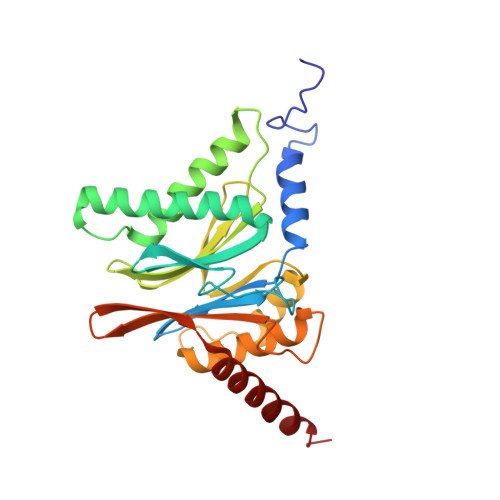
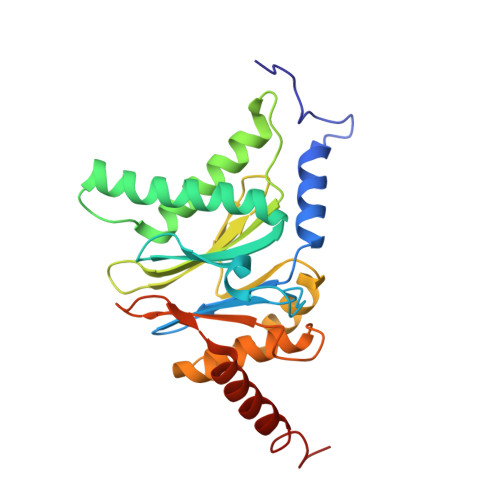
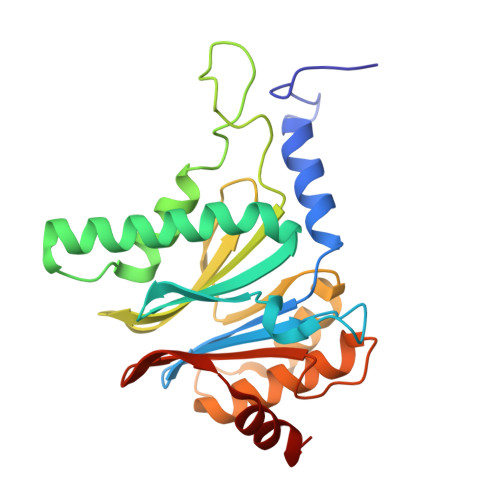
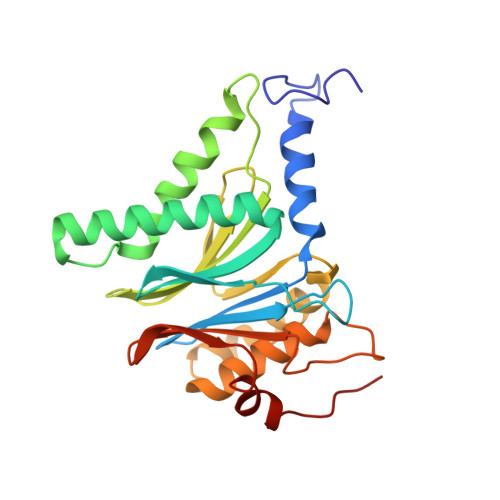
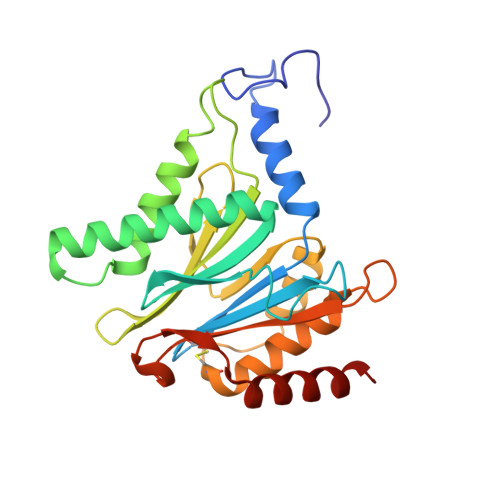
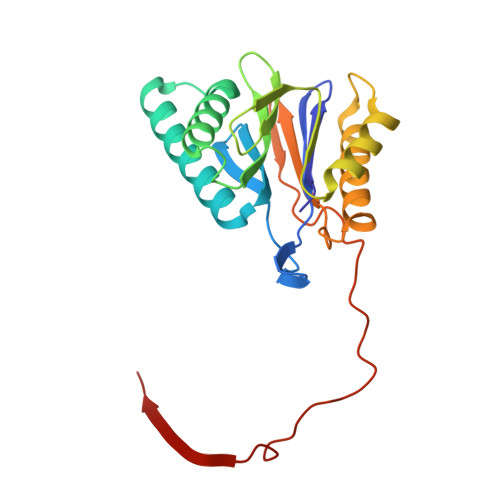
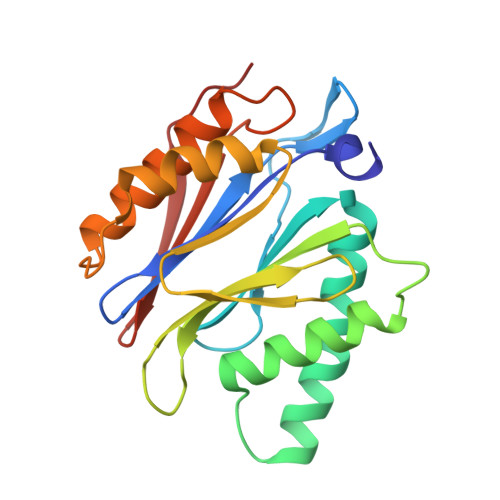
Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Harshbarger, W., Miller, C., Diedrich, C., Sacchettini, J.(2015) Structure 23: 418-424
- PubMed: 25599644
- DOI: https://doi.org/10.1016/j.str.2014.11.017
- Primary Citation of Related Structures:
4R3O, 4R67 - PubMed Abstract:
Proteasome inhibition is highly effective as a treatment for multiple myeloma, and recently carfilzomib was granted US FDA approval for the treatment of relapsed and refractory multiple myeloma. Here, we report the X-ray crystal structure of the human constitutive 20S proteasome with and without carfilzomib bound at 2.9 and 2.6 Å, respectively. Our data indicate that the S3 and S4 binding pockets play a pivotal role in carfilzomib's selectivity for chymotrypsin-like sites. Structural comparison with the mouse immunoproteasome crystal structure reveals amino acid substitutions that explain carfilzomib's slight preference for chymotrypsin-like subunits of constitutive proteasomes. In addition, comparison of the human proteasome:carfilzomib complex with the mouse proteasome:PR-957 complex reveals new details that explain why PR-957 is selective for immunoproteasomes. Together, the data presented here support the design of inhibitors for either constitutive or immunoproteasomes, with implications for the treatment of cancers as well as autoimmune and neurodegenerative diseases.
- Department of Chemistry, Texas A&M University, College Station, TX 77842-3012, USA.
Organizational Affiliation: