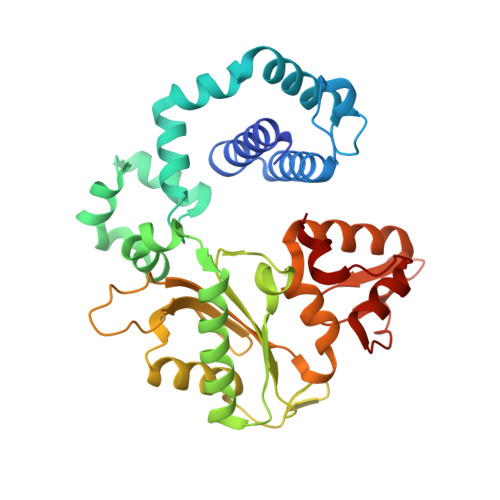
Substrate-induced DNA Polymerase beta Activation.
Beard, W.A., Shock, D.D., Batra, V.K., Prasad, R., Wilson, S.H.(2014) J Biological Chem 289: 31411-31422
- PubMed: 25261471
- DOI: https://doi.org/10.1074/jbc.M114.607432
- Primary Citation of Related Structures:
4R63, 4R64, 4R65, 4R66 - PubMed Abstract:
DNA polymerases and substrates undergo conformational changes upon forming protein-ligand complexes. These conformational adjustments can hasten or deter DNA synthesis and influence substrate discrimination. From structural comparison of binary DNA and ternary DNA-dNTP complexes of DNA polymerase β, several side chains have been implicated in facilitating formation of an active ternary complex poised for chemistry. Site-directed mutagenesis of these highly conserved residues (Asp-192, Arg-258, Phe-272, Glu-295, and Tyr-296) and kinetic characterization provides insight into the role these residues play during correct and incorrect insertion as well as their role in conformational activation. The catalytic efficiencies for correct nucleotide insertion for alanine mutants were wild type ∼ R258A > F272A ∼ Y296A > E295A > D192A. Because the efficiencies for incorrect insertion were affected to about the same extent for each mutant, the effects on fidelity were modest (<5-fold). The R258A mutant exhibited an increase in the single-turnover rate of correct nucleotide insertion. This suggests that the wild-type Arg-258 side chain generates a population of non-productive ternary complexes. Structures of binary and ternary substrate complexes of the R258A mutant and a mutant associated with gastric carcinomas, E295K, provide molecular insight into intermediate structural conformations not appreciated previously. Although the R258A mutant crystal structures were similar to wild-type enzyme, the open ternary complex structure of E295K indicates that Arg-258 stabilizes a non-productive conformation of the primer terminus that would decrease catalysis. Significantly, the open E295K ternary complex binds two metal ions indicating that metal binding cannot overcome the modified interactions that have interrupted the closure of the N-subdomain.
- From the Laboratory of Structural Biology, NIEHS, National Institutes of Health, Research Triangle Park, North Carolina 27709.
Organizational Affiliation: