Discovery of O-(3-carbamimidoylphenyl)-l-serine amides as matriptase inhibitors using a fragment-linking approach
Goswami, R., Wohlfahrt, G., Mukherjee, S., Ghadiyaram, C., Nagaraj, J., Satyam, L.K., Subbarao, K., Gopinath, S., Krishnamurthy, N.R., Subramanya, H.S., Ramachandra, M.(2015) Bioorg Med Chem Lett 25: 616-620
- PubMed: 25556099
- DOI: https://doi.org/10.1016/j.bmcl.2014.12.008
- Primary Citation of Related Structures:
4R0I - PubMed Abstract:
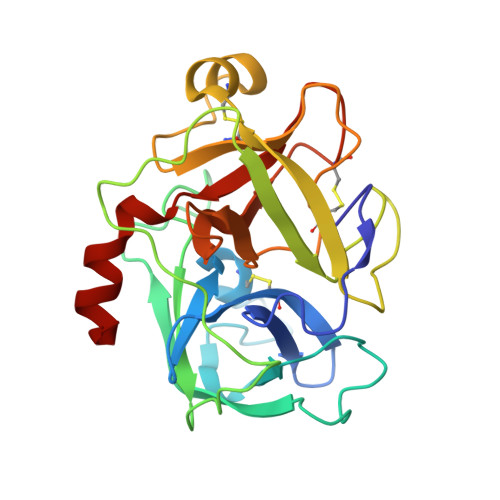
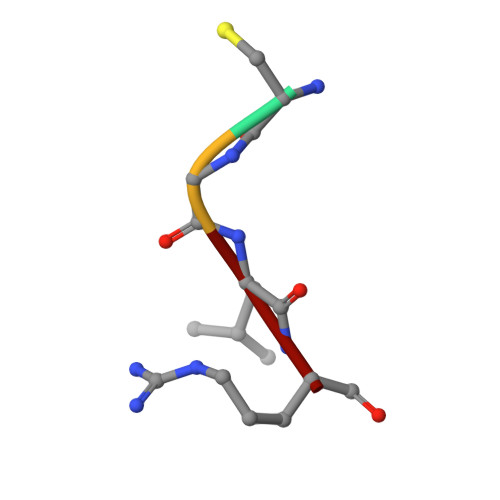
Matriptase is a cell-surface trypsin-like serine protease of epithelial origin, which cleaves and activates proteins including hepatocyte growth factor/scatter factor and proteases such as uPA, which are involved in the progression of various cancers. Here we report a fragment-linking approach, which led to the discovery of O-(3-carbamimidoylphenyl)-l-serine amides as potent matriptase inhibitors. The co-crystal structure of one of the potent inhibitors, 6 in complex with matriptase catalytic domain validated the working hypothesis guiding the development of this congeneric series and revealed the structural basis for matriptase inhibition. Replacement of a naphthyl group in 6 with 2,4,6-tri-isopropyl phenyl resulted in 10 with improved matriptase inhibition, which exhibited significant primary tumor growth inhibition in a mouse model of prostate cancer. Compounds such as 10, identified using a fragment-linking approach, can be explored further to understand the role of matriptase as a drug target in cancer and inflammation.
- Aurigene Discovery Technologies Limited, 39-40 KIADB Industrial Area, Electronic City Phase II, Bangalore 560 100, India.
Organizational Affiliation: