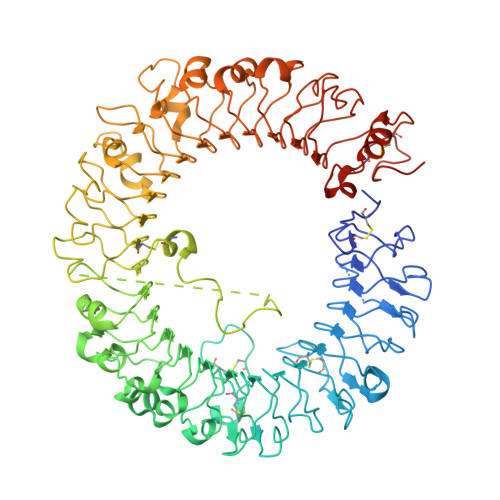
Toll-like receptor 8 senses degradation products of single-stranded RNA.
Tanji, H., Ohto, U., Shibata, T., Taoka, M., Yamauchi, Y., Isobe, T., Miyake, K., Shimizu, T.(2015) Nat Struct Mol Biol 22: 109-115
- PubMed: 25599397
- DOI: https://doi.org/10.1038/nsmb.2943
- Primary Citation of Related Structures:
4R07, 4R08, 4R09, 4R0A - PubMed Abstract:
Toll-like receptor 8 (TLR8) recognizes viral or bacterial single-stranded RNA (ssRNA) and activates innate immune systems. TLR8 is activated by uridine- and guanosine-rich ssRNA as well as by certain synthetic chemicals; however, the molecular basis for ssRNA recognition has remained unknown. In this study, to elucidate the recognition mechanism of ssRNA, we determined the crystal structures of human TLR8 in complex with ssRNA. TLR8 recognized two degradation products of ssRNA—uridine and a short oligonucleotide—at two distinct sites: uridine bound the site on the dimerization interface where small chemical ligands are recognized, whereas short oligonucleotides bound a newly identified site on the concave surface of the TLR8 horseshoe structure. Site-directed mutagenesis revealed that both binding sites were essential for activation of TLR8 by ssRNA. These results demonstrate that TLR8 is a sensor for both uridine and a short oligonucleotide derived from RNA.
- Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo, Japan.
Organizational Affiliation: