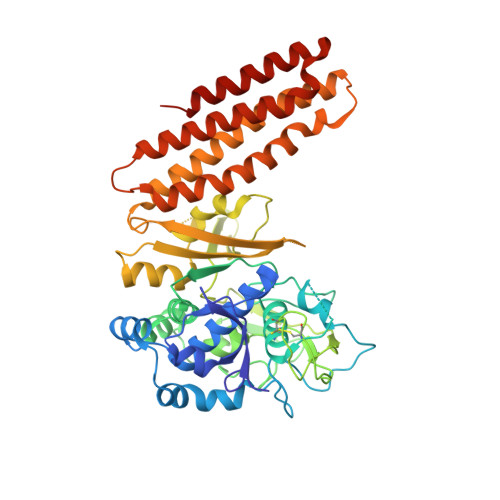
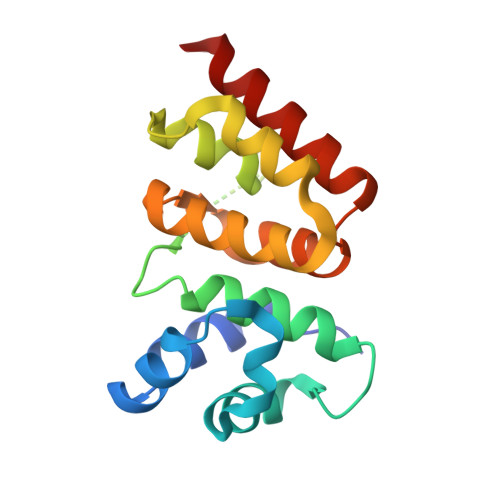
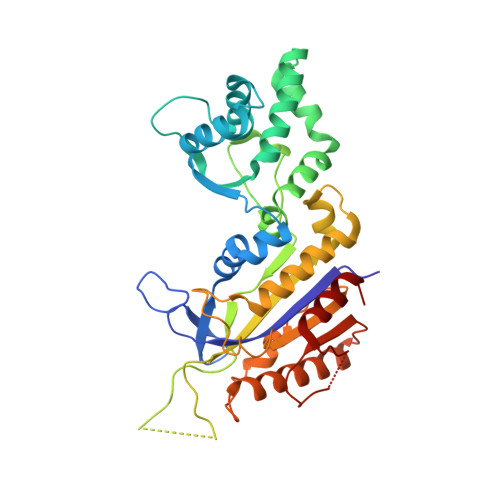
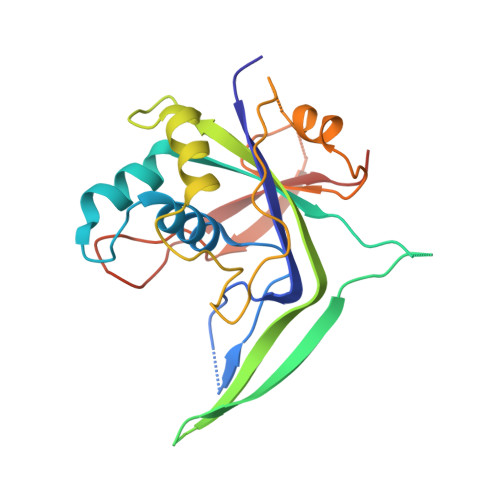
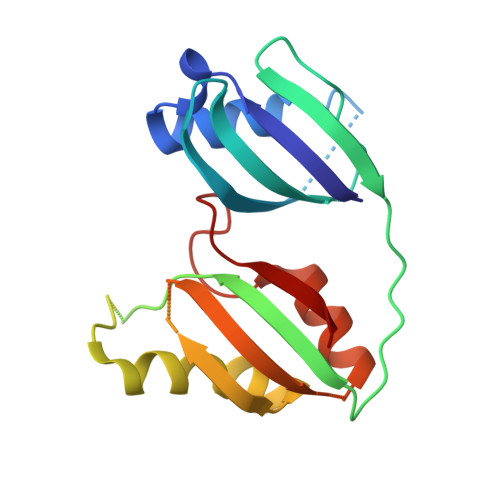
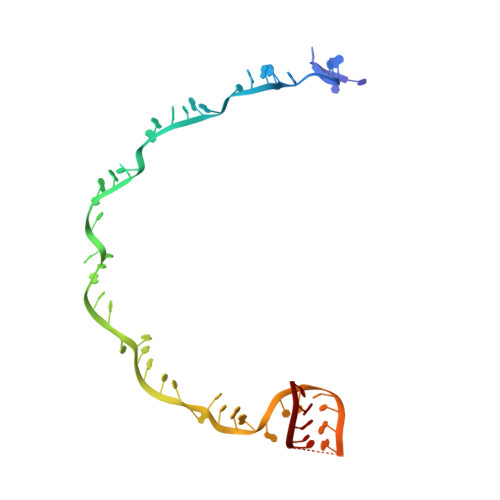
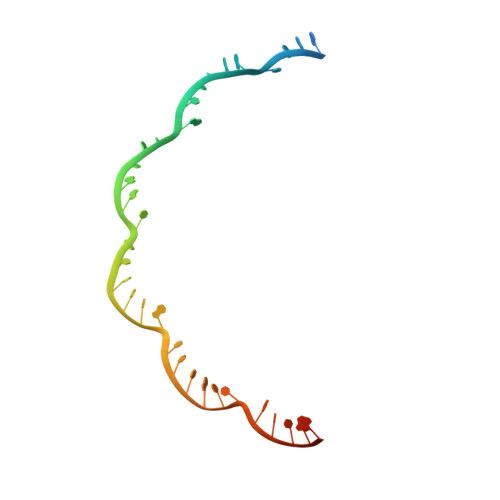
Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target.
Mulepati, S., Heroux, A., Bailey, S.(2014) Science 345: 1479-1484
- PubMed: 25123481
- DOI: https://doi.org/10.1126/science.1256996
- Primary Citation of Related Structures:
4QYZ - PubMed Abstract:
In prokaryotes, RNA derived from type I and type III CRISPR loci direct large ribonucleoprotein complexes to destroy invading bacteriophage and plasmids. In Escherichia coli, this 405-kilodalton complex is called Cascade. We report the crystal structure of Cascade bound to a single-stranded DNA (ssDNA) target at a resolution of 3.03 angstroms. The structure reveals that the CRISPR RNA and target strands do not form a double helix but instead adopt an underwound ribbon-like structure. This noncanonical structure is facilitated by rotation of every sixth nucleotide out of the RNA-DNA hybrid and is stabilized by the highly interlocked organization of protein subunits. These studies provide insight into both the assembly and the activity of this complex and suggest a mechanism to enforce fidelity of target binding.
- Department of Biochemistry and Molecular Biology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD 21205, USA.
Organizational Affiliation: