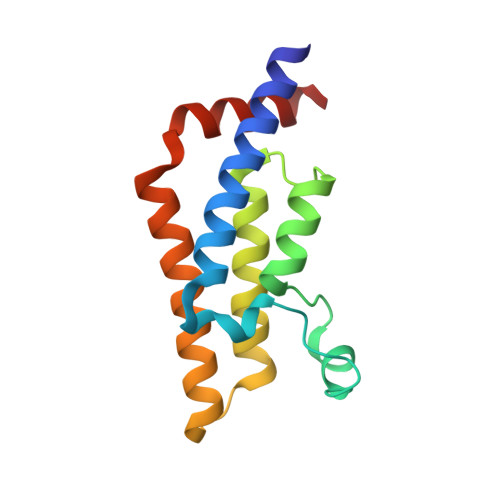
Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells.
Morozumi, Y., Boussouar, F., Tan, M., Chaikuad, A., Jamshidikia, M., Colak, G., He, H., Nie, L., Petosa, C., de Dieuleveult, M., Curtet, S., Vitte, A.L., Rabatel, C., Debernardi, A., Cosset, F.L., Verhoeyen, E., Emadali, A., Schweifer, N., Gianni, D., Gut, M., Guardiola, P., Rousseaux, S., Gerard, M., Knapp, S., Zhao, Y., Khochbin, S.(2016) J Mol Cell Biol 8: 349-362
- PubMed: 26459632
- DOI: https://doi.org/10.1093/jmcb/mjv060
- Primary Citation of Related Structures:
4QUT, 4QUU - PubMed Abstract:
Although the conserved AAA ATPase and bromodomain factor, ATAD2, has been described as a transcriptional co-activator upregulated in many cancers, its function remains poorly understood. Here, using a combination of ChIP-seq, ChIP-proteomics, and RNA-seq experiments in embryonic stem cells where Atad2 is normally highly expressed, we found that Atad2 is an abundant nucleosome-bound protein present on active genes, associated with chromatin remodelling, DNA replication, and DNA repair factors. A structural analysis of its bromodomain and subsequent investigations demonstrate that histone acetylation guides ATAD2 to chromatin, resulting in an overall increase of chromatin accessibility and histone dynamics, which is required for the proper activity of the highly expressed gene fraction of the genome. While in exponentially growing cells Atad2 appears dispensable for cell growth, in differentiating ES cells Atad2 becomes critical in sustaining specific gene expression programmes, controlling proliferation and differentiation. Altogether, this work defines Atad2 as a facilitator of general chromatin-templated activities such as transcription.
- INSERM, U823; Université Grenoble Alpes; Institut Albert Bonniot Grenoble, F-38700 Grenoble, France.
Organizational Affiliation: