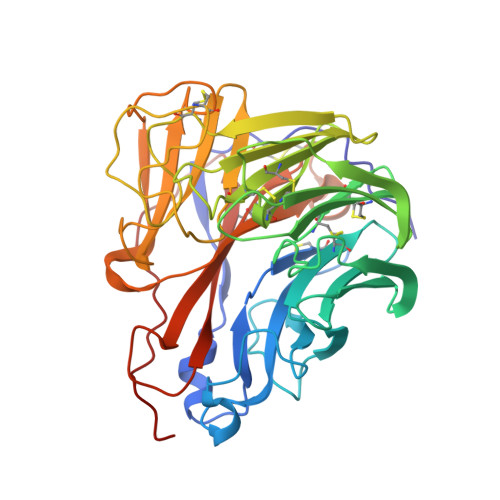
Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers.
Wan, H., Yang, H., Shore, D.A., Garten, R.J., Couzens, L., Gao, J., Jiang, L., Carney, P.J., Villanueva, J., Stevens, J., Eichelberger, M.C.(2015) Nat Commun 6: 6114-6114
- PubMed: 25668439
- DOI: https://doi.org/10.1038/ncomms7114
- Primary Citation of Related Structures:
4QNP - PubMed Abstract:
A(H1N1)pdm09 influenza A viruses predominated in the 2013-2014 USA influenza season, and although most of these viruses remain sensitive to Food and Drug Administration-approved neuraminidase (NA) inhibitors, alternative therapies are needed. Here we show that monoclonal antibody CD6, selected for binding to the NA of the prototypic A(H1N1)pdm09 virus, A/California/07/2009, protects mice against lethal virus challenge. The crystal structure of NA in complex with CD6 Fab reveals a unique epitope, where the heavy-chain complementarity determining regions (HCDRs) 1 and 2 bind one NA monomer, the light-chain CDR2 binds the neighbouring monomer, whereas HCDR3 interacts with both monomers. This 30-amino-acid epitope spans the lateral face of an NA dimer and is conserved among circulating A(H1N1)pdm09 viruses. These results suggest that the large, lateral CD6 epitope may be an effective target of antibodies selected for development as therapeutic agents against circulating H1N1 influenza viruses.
- Division of Viral Products, Center for Biologics Evaluation and Research, Food and Drug Administration, 10903 New Hampshire Avenue, Silver Spring, Maryland 20993, USA.
Organizational Affiliation: