Crystal structure of euryarchaeal RNA polymerase and insight into the evolution of RNA polymerase II structure
Murakami, K.S.(2014) Nat Commun
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2014) Nat Commun
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase, subunit E' | 190 | Thermococcus kodakarensis KOD1 | Mutation(s): 0 Gene Names: Rpo7, TK1699 EC: 2.7.7.6 | 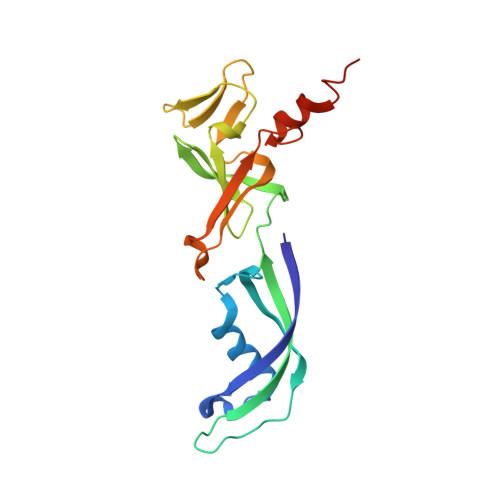 | |
UniProt | |||||
Find proteins for Q5JIY4 (Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1)) Explore Q5JIY4 Go to UniProtKB: Q5JIY4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5JIY4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase, subunit F | 122 | Thermococcus kodakarensis KOD1 | Mutation(s): 0 Gene Names: Rpo4, TK0901 EC: 2.7.7.6 | 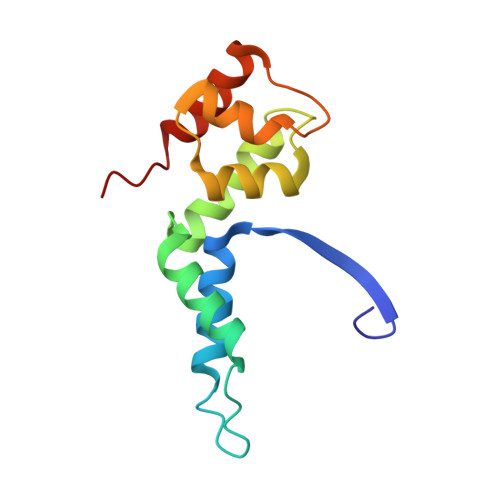 | |
UniProt | |||||
Find proteins for Q5JI52 (Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1)) Explore Q5JI52 Go to UniProtKB: Q5JI52 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5JI52 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.465 | α = 90 |
| b = 80.942 | β = 90 |
| c = 97.413 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Crystal | data collection |
| PHENIX | model building |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |