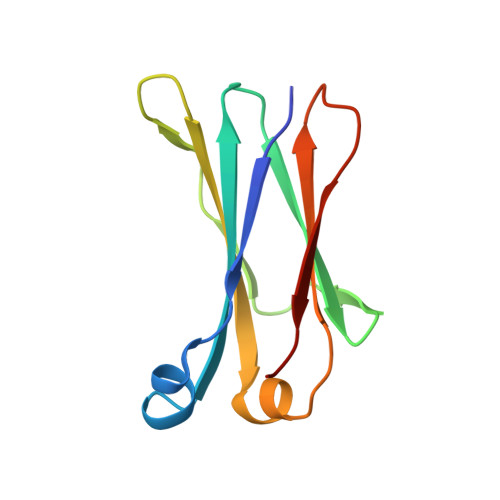
The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Feige, M.J., Grawert, M.A., Marcinowski, M., Hennig, J., Behnke, J., Auslander, D., Herold, E.M., Peschek, J., Castro, C.D., Flajnik, M., Hendershot, L.M., Sattler, M., Groll, M., Buchner, J.(2014) Proc Natl Acad Sci U S A 111: 8155-8160
- PubMed: 24830426
- DOI: https://doi.org/10.1073/pnas.1321502111
- Primary Citation of Related Structures:
2MKL, 4Q97, 4Q9B, 4Q9C - PubMed Abstract:
Sharks and other cartilaginous fish are the phylogenetically oldest living organisms that rely on antibodies as part of their adaptive immune system. They produce the immunoglobulin new antigen receptor (IgNAR), a homodimeric heavy chain-only antibody, as a major part of their humoral adaptive immune response. Here, we report the atomic resolution structure of the IgNAR constant domains and a structural model of this heavy chain-only antibody. We find that despite low sequence conservation, the basic Ig fold of modern antibodies is already present in the evolutionary ancient shark IgNAR domains, highlighting key structural determinants of the ubiquitous Ig fold. In contrast, structural differences between human and shark antibody domains explain the high stability of several IgNAR domains and allowed us to engineer human antibodies for increased stability and secretion efficiency. We identified two constant domains, C1 and C3, that act as dimerization modules within IgNAR. Together with the individual domain structures and small-angle X-ray scattering, this allowed us to develop a structural model of the complete IgNAR molecule. Its constant region exhibits an elongated shape with flexibility and a characteristic kink in the middle. Despite the lack of a canonical hinge region, the variable domains are spaced appropriately wide for binding to multiple antigens. Thus, the shark IgNAR domains already display the well-known Ig fold, but apart from that, this heavy chain-only antibody employs unique ways for dimerization and positioning of functional modules.
- Department of Tumor Cell Biology, St. Jude Children's Research Hospital, Memphis, TN 38105; matthias.feige@stjude.org.
Organizational Affiliation: